Hello all,
I am doing a Bayesian calibration (updating) of model parameters using UqLab.
I am applying the updating on a simple dynamic system with experimental data (eigenvalues) under controlled parameter uncertainty - since I know the uncertainty exactly, the posterior should replicate the known uncertainty very closely if my model is good.
My problem seems to be in understanding the meaning of the discrepancy and how it is used in UqLab.
As I understand it, the discrepancy is the difference between my experimental observation and my model prediction before the updating. The posterior predictive model should be very close to the experimental data - with very small discrepancy.
I implemented the discrepancy as explained in section 2.2.6.1 of the user manual:
DiscrepancyOpts.Type = 'Gaussian';
DiscrepancyOpts.Parameters = [5.2696e+03, 1.9387e+05, 4.7241e+05];
However, the updated parameters are not as expected (the mean is correct, but the variance is too small – I should get an STD around 0.0145):
%------------------- Posterior Marginals
------------------------------------------------------------
| Parameter | Mean | Std | (0.025-0.97) Quant. | Type |
------------------------------------------------------------
| RL12 | 0.05 | 0.0017 | (0.046 - 0.053) | Model |
| RL23 | 0.053 | 0.0014 | (0.05 - 0.056) | Model |
------------------------------------------------------------
%------------------- Point estimate
--------------------------------------
| Parameter | Mean | Parameter Type |
--------------------------------------
| RL12 | 0.05 | Model |
| RL23 | 0.053 | Model |
--------------------------------------
%------------------- Correlation matrix (model parameters)
-----------------------------
| | RL12 RL23 |
-----------------------------
| RL12 | 1 -0.078 |
| RL23 | -0.078 1 |
-----------------------------
Here are figures showing the updated parameter distributions and also the prior predictive and posterior predictive eigenvalue distributions.
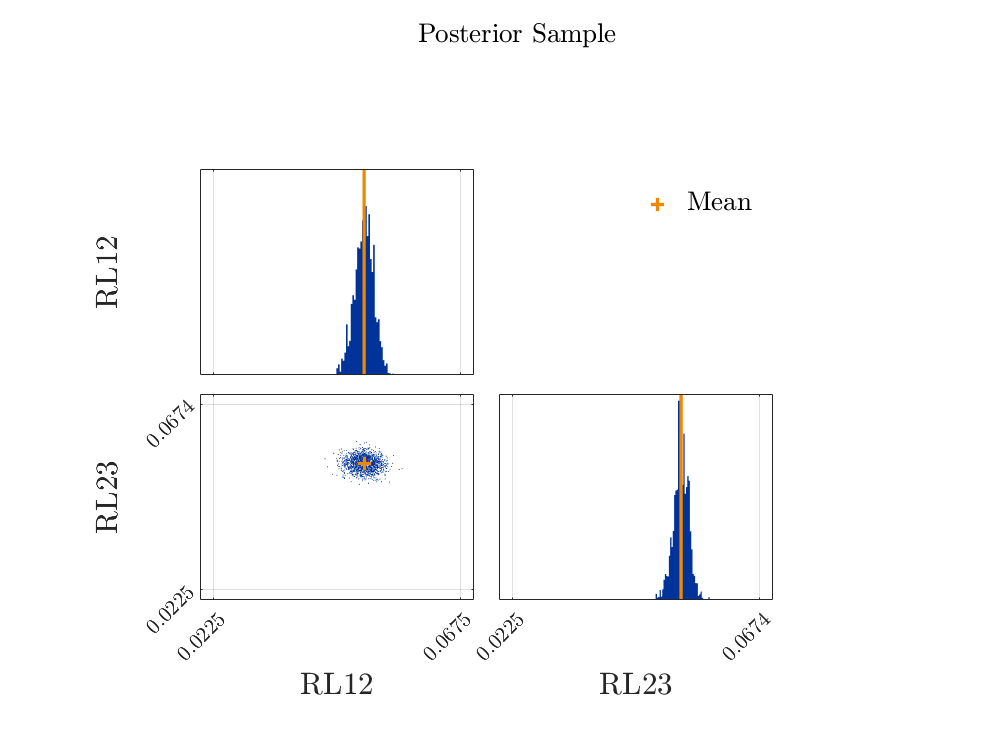
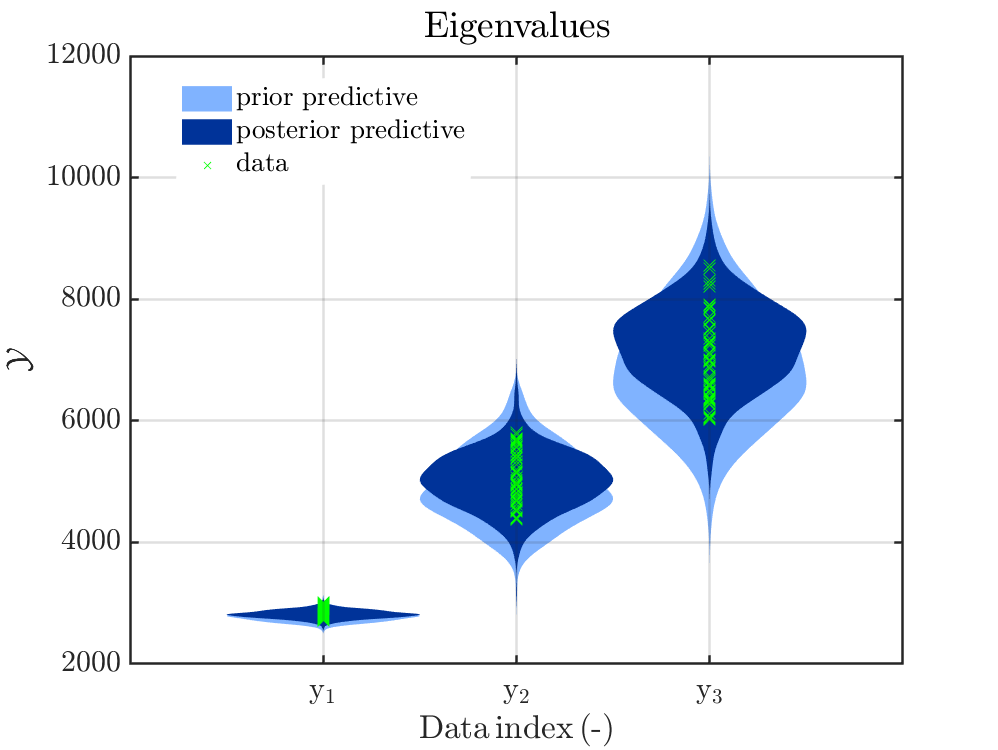
The posterior predictive eigenvalue distributions seem to be correct, but the updated parameter distributions are too narrow. If, using the updated parameter distributions, I regenerate the eigenvalue distributions, then they are too narrow and do not agree with those in the figure. It seems that UqLab adds something to the predictive posterior (eigenvalue) distributions. Could it be that the prior discrepancy was added to the posterior eigenvalue prediction?
I am almost sure the problem is in how I calculated the discrepancy model from the manuals - it is not clear to me how to do that. Someone can help me please?
I also assumed the discrepancy unknown (section 2.2.6.2 of the manual). However, the results are close to the ones described above.