Dear Paul-Remo,
thanks for your answer. I am still on my way of understanding and testing
your suggestion, but I have already a small remark and a big one:
1.) Since consistency checks have quite often saved my day by pointing out
that I made some typo etc. I would prefer to keep the consistency check
in general, but to allow some override by adding a list in data.MOMapNotUnique
of those models to point out that I really want to reuse at least one of
outputs of the model. Hence, I replaced in uq_initialize_uq_inversion
the lines 175-177 by
if length(unique(CurrModelOMap)) ~= length(CurrModelOMap)
if ~isfield(Opt.Value(1),'MOMapNotUnique')
fprintf('\n Problem with model \%g\n',ii);
error('The supplied MOMap does not uniquely address every model output and no exception list in MOMapNotUnique is provided')
elseif ~ismember(ii,Opt.Value(1).MOMapNotUnique)
fprintf('\n Problem with model \%g:\n',ii);
error('The supplied MOMap does not uniquely address one model output and the model is not listed in the exception list in MOMapNotUnique')
\% else case would correspond to the situation that the supplied
\% MOMap does not uniquely address every model output but that is
\% no error and therefroe announced by inserting the model
\% number in the exception list in MOMapNotUnique
\% BUT, one may have to add a further check to ensure that
\% that no output does appear in several data groups to ensure
\% the the outout has one unique discrepancy assigned to it,
end
elseif isfield(Opt.Value(1),'MOMapNotUnique') && ismember(ii,Opt.Value(1).MOMapNotUnique)
fprintf('\n Problem with model \%g:\n',ii);
error('The supplied MOMap does uniquely address all outputs for this model but the model is listed in MOMapNotUnique')
end
- To understand the results of my tests I needed to learn more on the components of
myBayesianAnalysis.Results.PostProc
that are not discussed in the manual. Hence, I examined the code in uq_postProcessInversion.m and realized that deactivating the consistency check may have side effects that can be quite surprising, at least according to my humble opinion:
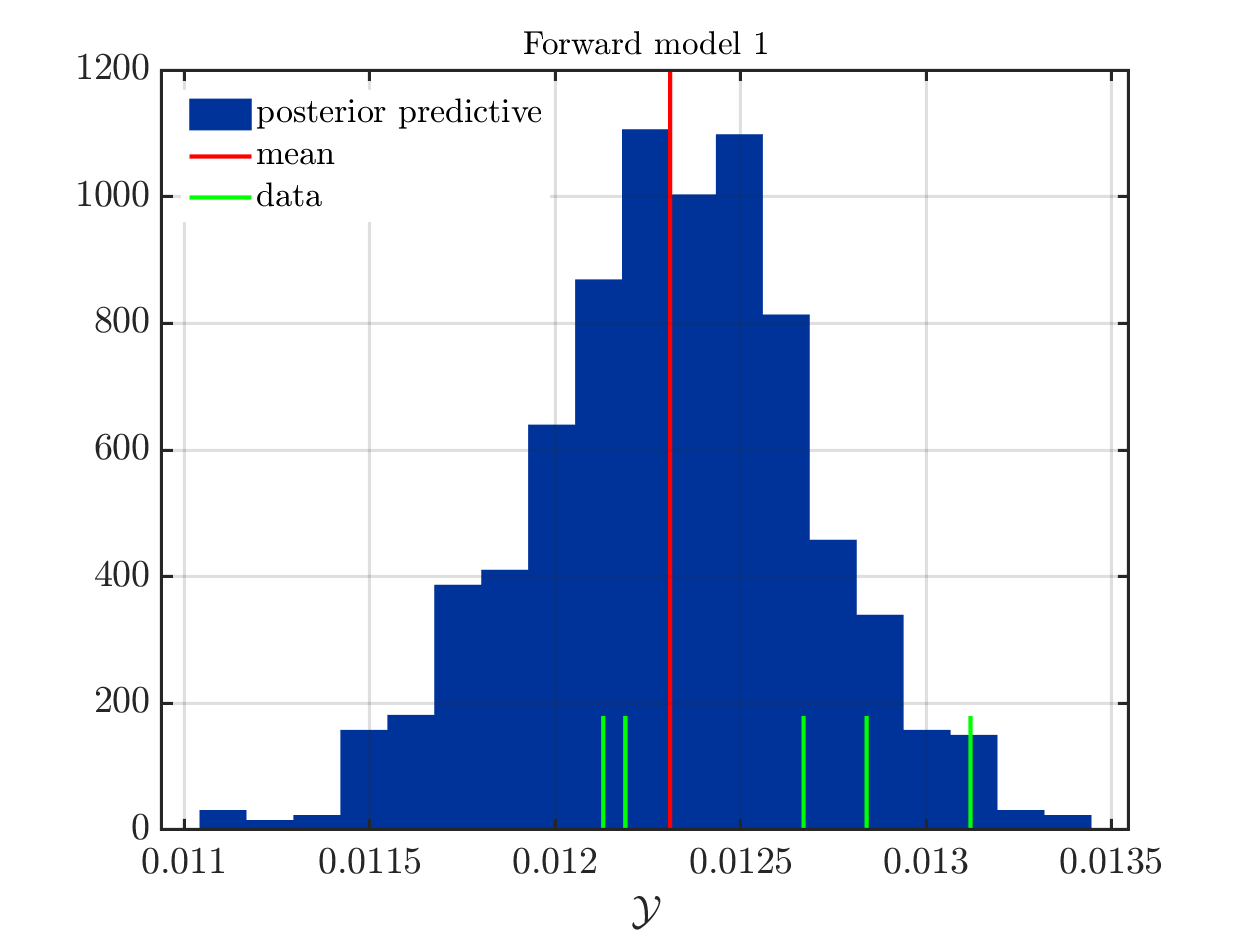
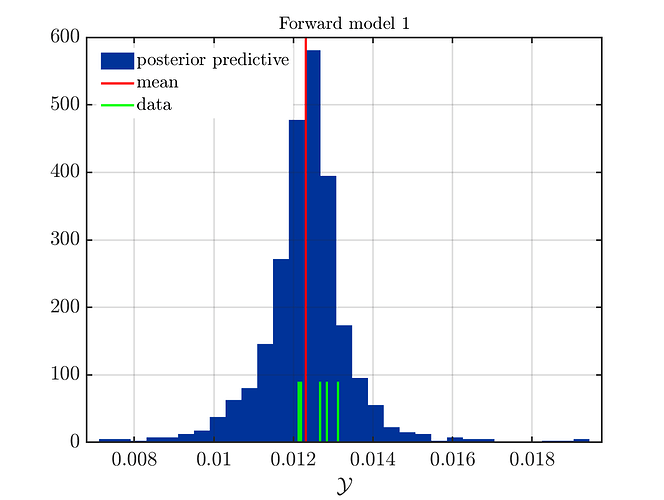
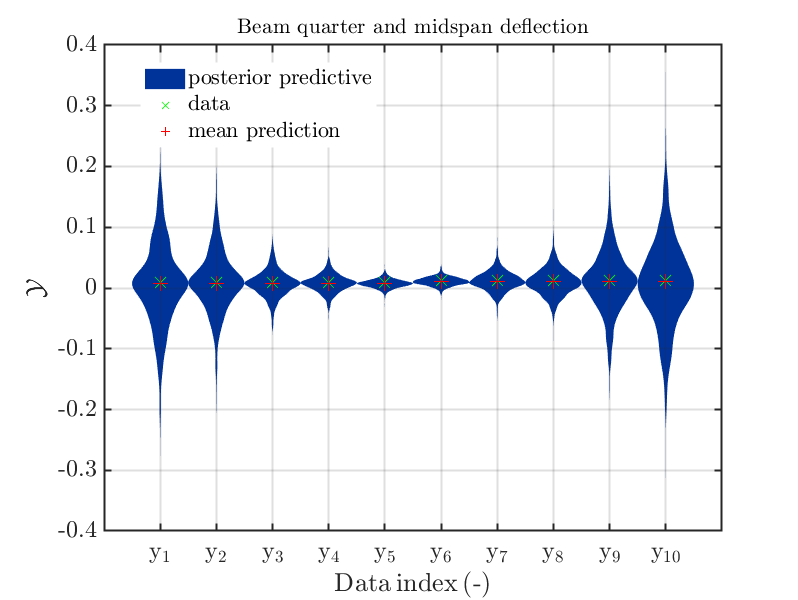
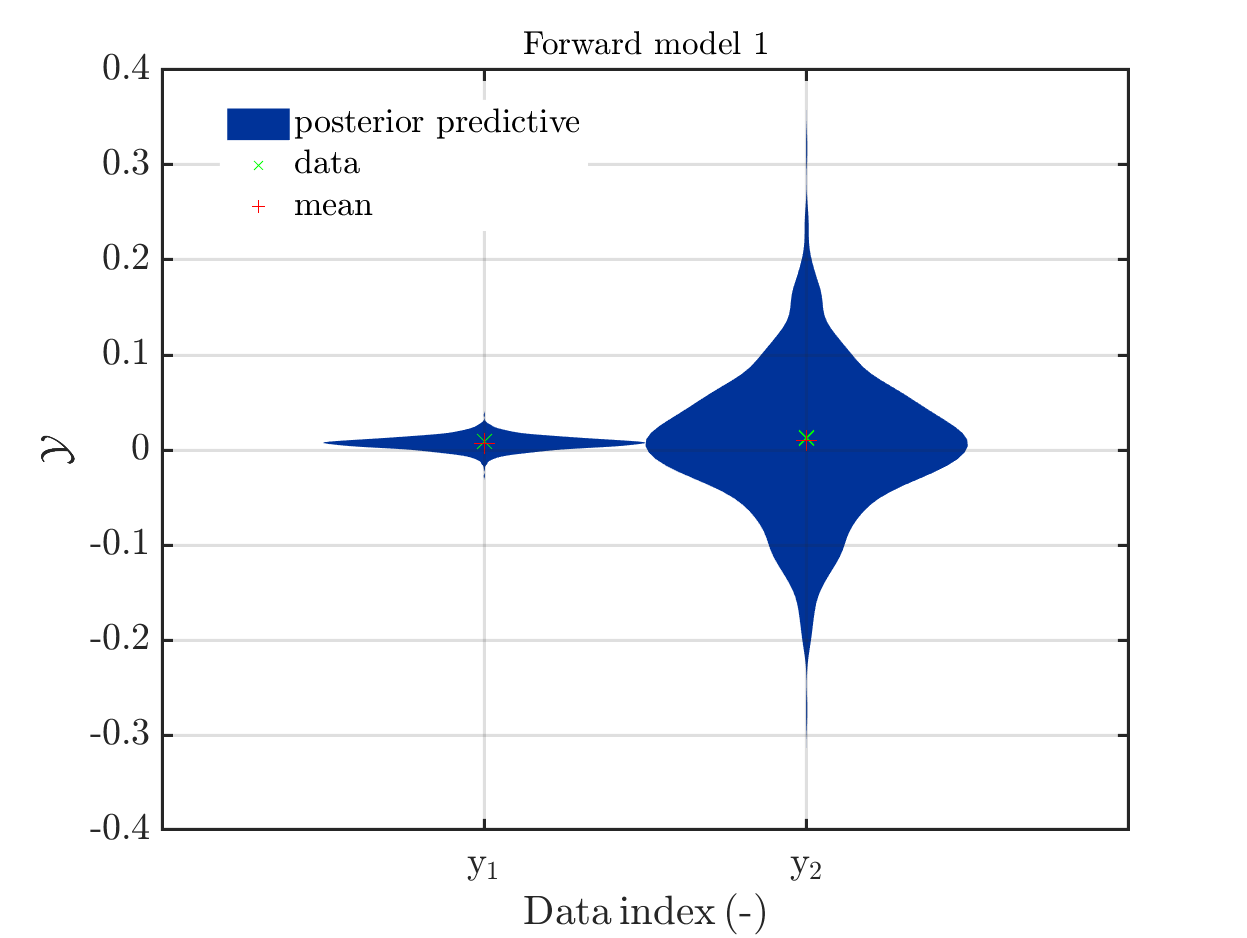
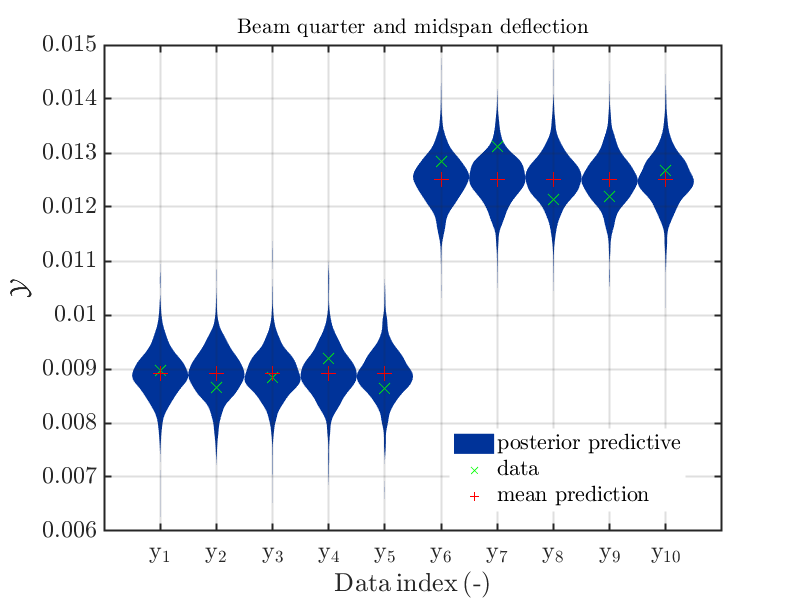
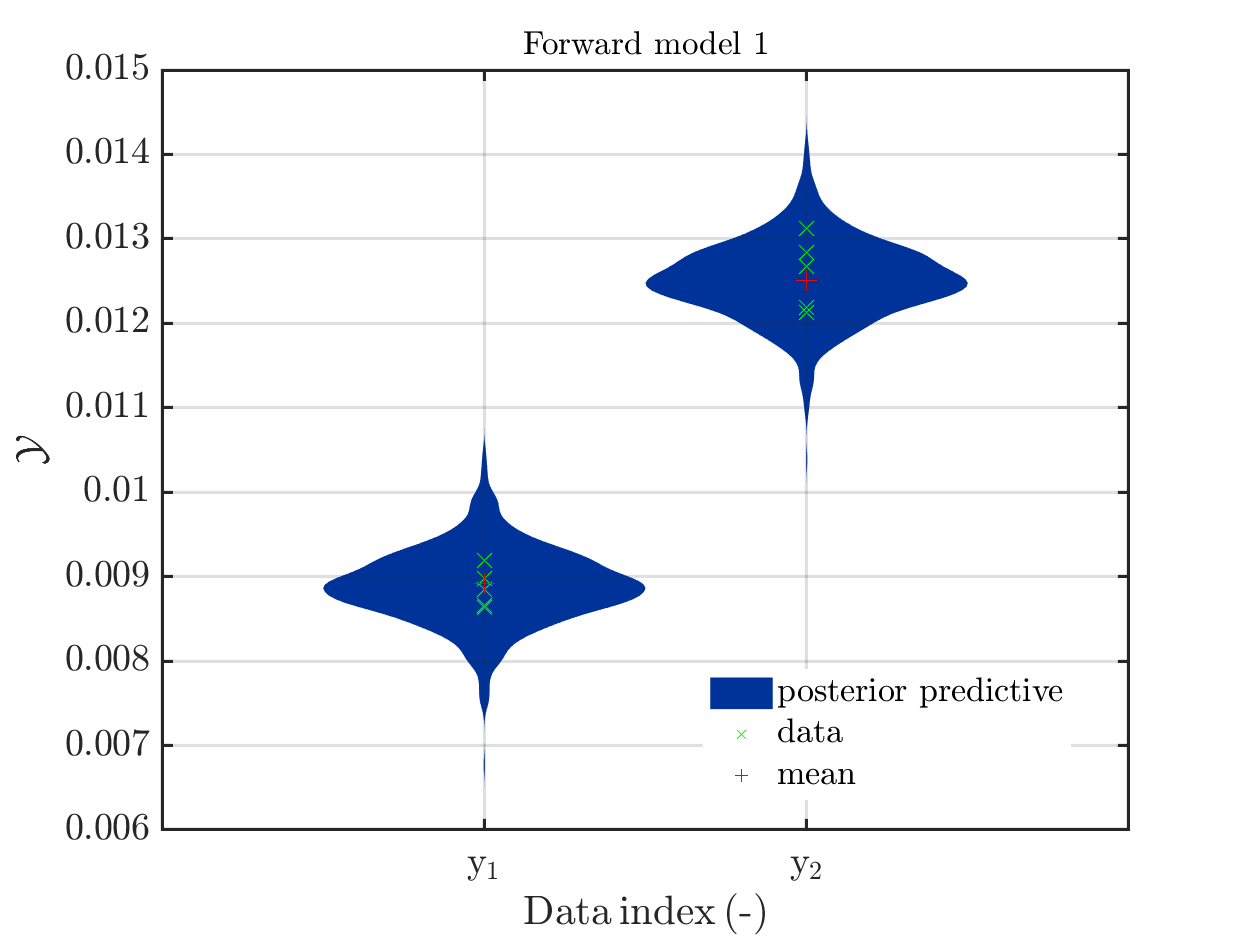
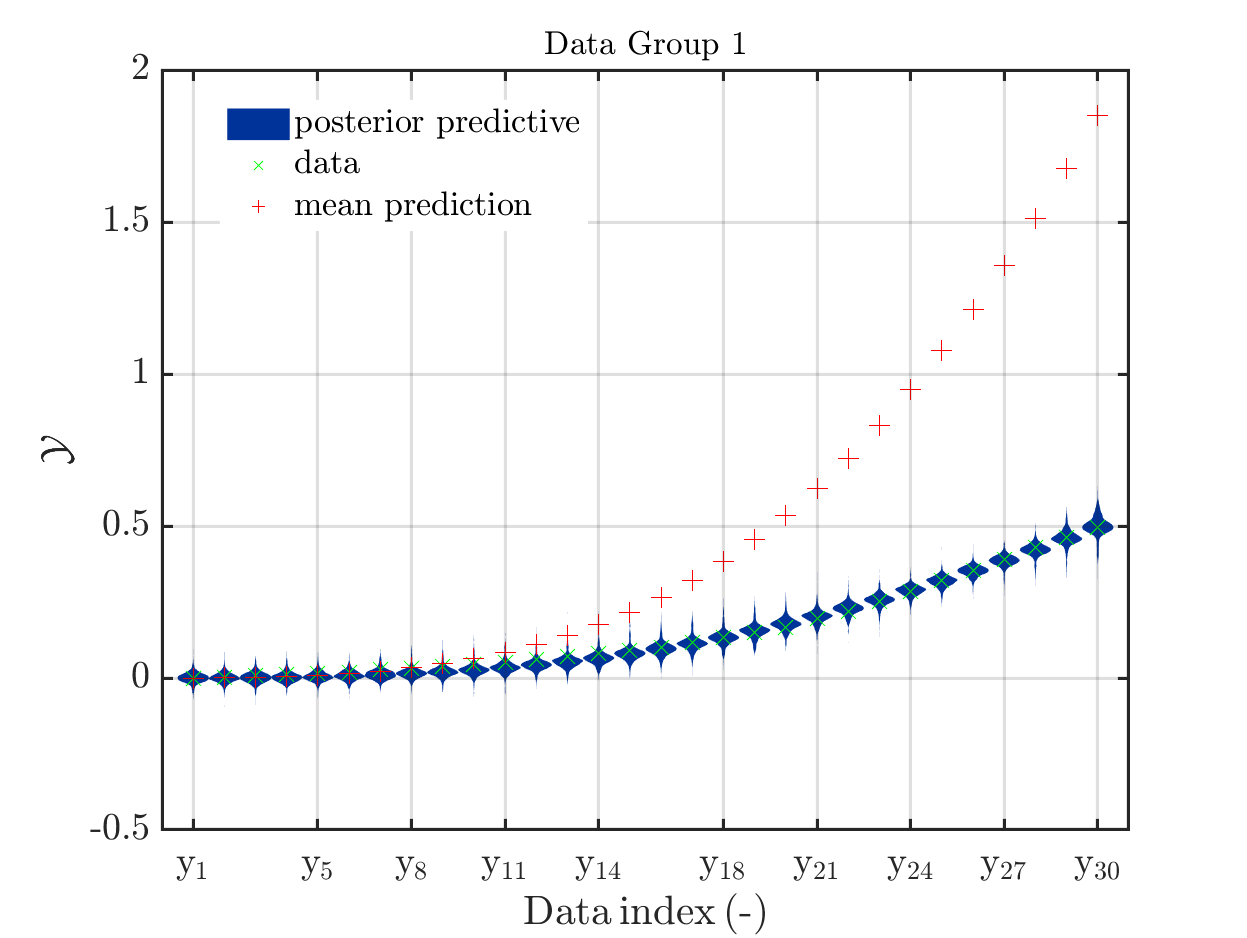
If one is using the same model output in two discrepancy data groups, one can change the resulting posterior predictive distributions plot by exchanging the groups, see SCR_DataGroup.m (2.5 KB) and SCR_DataGroupExc.m (2.6 KB) leading to the posterior predictive distributions plots in
and in
, respectively.
This is a emulation of the situation that a UQLab-user has two sets of measurements for the same model, one with many measurements and some discrepancy and a second one with less measurements but also a smaller discrepancy.
Using two discrepancy data groups with two different UQLab models, both using the same function, the UQLab-user can identity the model parameter and can get two separate scatter plots. To get one combined scatter plot the UQLab-user may try to use the same UQLab model in both discrepancy data groups since she/he exepects that the created posterior predictive sample points used for the scatter plot, i.e.the contents of
myBayesianAnalysis.Results.PostProc.PostPred.model(mm).postPredRuns
are somehow randomly choosen from all predictive sample values computed in both (all) data groups.
(It holds for
myBayesianAnalysis.Results.PostProc.PostPred.model(mm).data
that the data sets provided in the different discrepancy data groups are connected
in one data set.)
But, this is not the case: Considering line the loop in the lines 520 to 576 in uq_postProcessInversion.m one realizes that for every model mm and every output number modelOutputNr it holds that
model(mm).postPredRuns(:,modeCompNr)
is equal to the predictive sample computed in the last discrepancy data groups containing the output modelOutputNr of model mm.
I think this is a problem one has to be aware of and may require to either disallow to use one model output in several discrepancy data groups or to change the loop discussed above to do same sampling between the different predictive sample values for the considered model output.
But, before chancing the loop one would need to fix a well defined way to choose predictive sample values for a model output if there are several data groups each one generating several predictive sample values, such that the loop in lines 563 - 575 starts with a field postPredRunsCurr, such for each index kk it holds that postPredRunsCurr(kk,: ) contains several several predictive sample values for the considered model output.
But, this is no direct problem for me since currently I plan to use only one discrepancy data group and will now continue my testing and will start to connect my problem to UQLab in the near future.
Best regards
Olaf