Hello,
I observed some unexpected and some undocumented behavior of
uq_postProcessInversion.m, that may require some changes in the code
and in the manual UQLab - Manual for Bayesian inversion
1) After reading the code in uq_postProcessInversion.m I started to believe
that applying ‘uq_postProcessInversion’ several times to the same
uq_analysis variable, which will be denoted by ``myBayesianAnalysis’’
in the following, with different options will produce finally the same output as applying uq_postProcessInversion to myBayesianAnalysis only one time with the last set of options.
If this assumption is correct, I suggest to point out this explicitly in the documentation.
An improved version of the following suggestion can be found below in the my reply to my request. Olaf
For example, you could write that the replacing the command
uq_postProcessInversion(myBayesianAnalysis,... 'badChains', [1 5], 'pointEstimate', 'MAP')
(see page 55 in the manual) by
uq_postProcessInversion(myBayesianAnalysis,... 'badChains', [1 5]) uq_postProcessInversion(myBayesianAnalysis,... 'pointEstimate', 'MAP')
will generate the following result:
- During the first call to uq_postProcessInversion the data in BayesianAnalysis.Results.PostPro are generated by “removing” the samples points generated by the first and fifth chains from the sample.
- But, during the second call to uq_postProcessInversion, the data in BayesianAnalysis.Results.PostPro are not directly updated but recreated using the information in myBayesianAnalysis.Results.Sample and in BayesianAnalysis.Results.ForwardModel, i.e. the data “removed” during
the first call are used.
End of suggestion that is improved in next post.
2.) If my assumption on the default esults of multiple calls to uq_postProcessInversion to myBayesianAnalysis is correct then there seems to be an exception if the option ‘priorPredictive’ is involved.
Testing and investigating the code I deduced: When uq_postProcessInversion
is called with the value provied for ‘priorPredictive’ being a positive integer,
the resulting samples from the prior distribution and from the prior predictive distribution are stored in myBayesianAnalysis.Results.PostProc.PriorPredSample.
But, if in the following uq_postProcessInversion is applied to myBayesianAnalysis without the ‘priorPredictive’ option, myBayesianAnalysis.Results.PostProc.PriorPredSample is not changed, i.e.
the resulting samples from the prior distribution and from the prior
predictive distribution are further aviable and can be used in the following.
Since creating this samples had required further function evaluations
and the sample seem to be independent of the values for badChains or bornIn, it can be considered as a feature if the samples are not automatically removed. But this is seems to be an exception of the normal behavior of uq_postProcessInversion, see also the discussion above. Therefore, I suggest be discuss this behavior in the manual.
3.) Moreover, this behavior can be quite irritating if one is trying to change the plots generated by uq_display(myBayesianAnalysis) by getting rid of these samples.
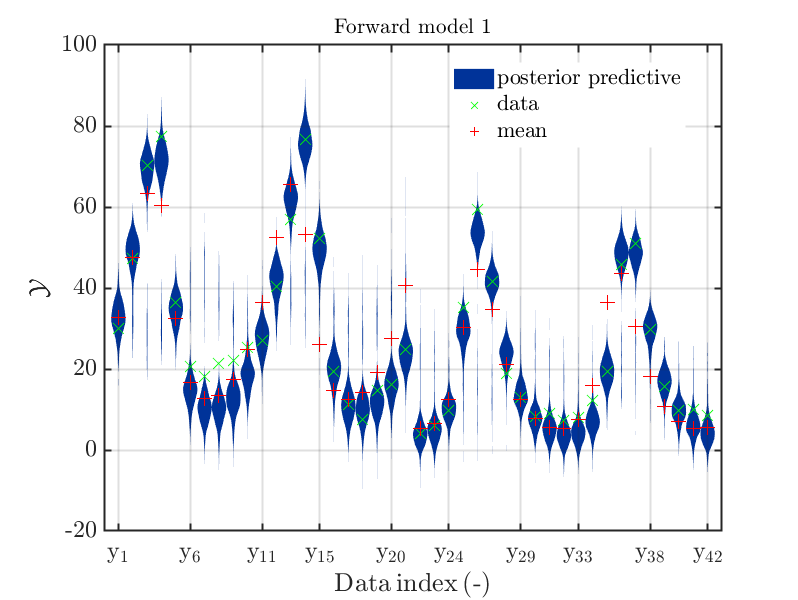
For example. after evaluating the model 1 plot generated by
uq_Example_Inversion_04_PredPrey.m, see the picture at the end of the example
INVERSION: PREDATOR-PREY MODEL CALIBRATION on the UQLab-Server:
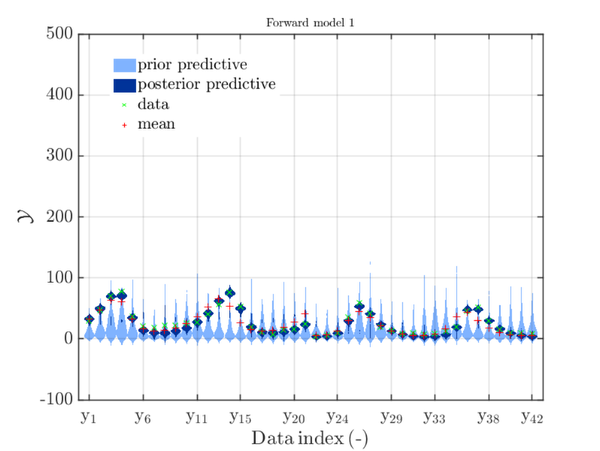
one may be interested to investigate this plot without the priori density distribution, see
And I like to able to do this without having to call uq_createAnalysis again. As pointed out above, one can not get rid of the samples for the priori predictive distribution by applying badChainsIndex to myBayesianAnalysis without using the priorPredictive.
Trying to get rid of the samples by using
uq_postProcessInversion(myBayesianAnalysis,…
‘badChains’, badChainsIndex,…
‘priorPredictive’, 0, …
‘posteriorPredictive’, 1000);
generated the error
Index exceeds matrix dimensions.
Error in uq_postProcessInversion (line 436)
priorRunsCurr(:,MIndex) = model(mm).priorRuns(:,OMapCurr);
Okay, this is somehow strange, since according to the documentation, 0 is the default value for ‘priorPredictive’, such that using this value should in my humble opinion not generate an error. In view of the code in uq_postProcessInversion one realizes that if the priorPredictive option is missing, the flag priorPredictive_flag is false and therefore some parts of the code are ignored. But, if for priorPredictive the value 0 is given, then it follows that the flag priorPredictive_flag is true and the value of nPriorPredSamples is 0, which seems not to create prolbems in the parts of the code that are not ignored since the flag is true.
Hence, I suggest to change the code of uq_postProcessInversion such that using the option ‘priorPredictive’ with value 0 creates the same result as not using the option 'priorPredictiv at all.
Even if one can find a way do get rid of the samples for the prior predictive distribution by a direct change of the contents of myBayesianAnalysis, I think that uq_postProcessInversion should provide a method to get rid of them.
For example, using a negative number for the option ‘priorPredictive’ could
a good way to activate this feature without changing the result of existing correct code.
4.) Moreover, concerning the documentation of uq_postProcessInversion: in Section 3.3 of the manual I suggest to point there that (starting with UQLab relase 1.3.0), this function is called with its default options at the end of the evaluation of uq_createAnalysis and maybe also of some other ULQllab function to automatically post-process the data, to generate a sample by any MCMC sampler and to create contents in ``myBayesianAnalysis.Results,PostProc. ‘’
Yes, this information can already be found in the release notes and in the comments in Section 6.3 of uq_Example_Inversion_03_Hydro.m but it would be nice if this would also be pointed out in the manual.
5) I could also be grateful, if the contents of myBayesianAnalysis.Results,PostProc
created by uq_postProcessInversion would be discussed in the manual.
Okay, I think that this text already to long.
Greetings
Olaf