Bayesian inference in UQLab: suggestions for improving the plot functions
and the corresponding UQLab User Manual
Dear @paulremo, dear all
after starting to work with UQLab, performing some Bayesian inference computations, examining the program code of UQLab, and pointing out some errors in the code, I would like to suggest some changes in the graphic output and in the UQLab User Manual for Bayesian inference from a users point of view:
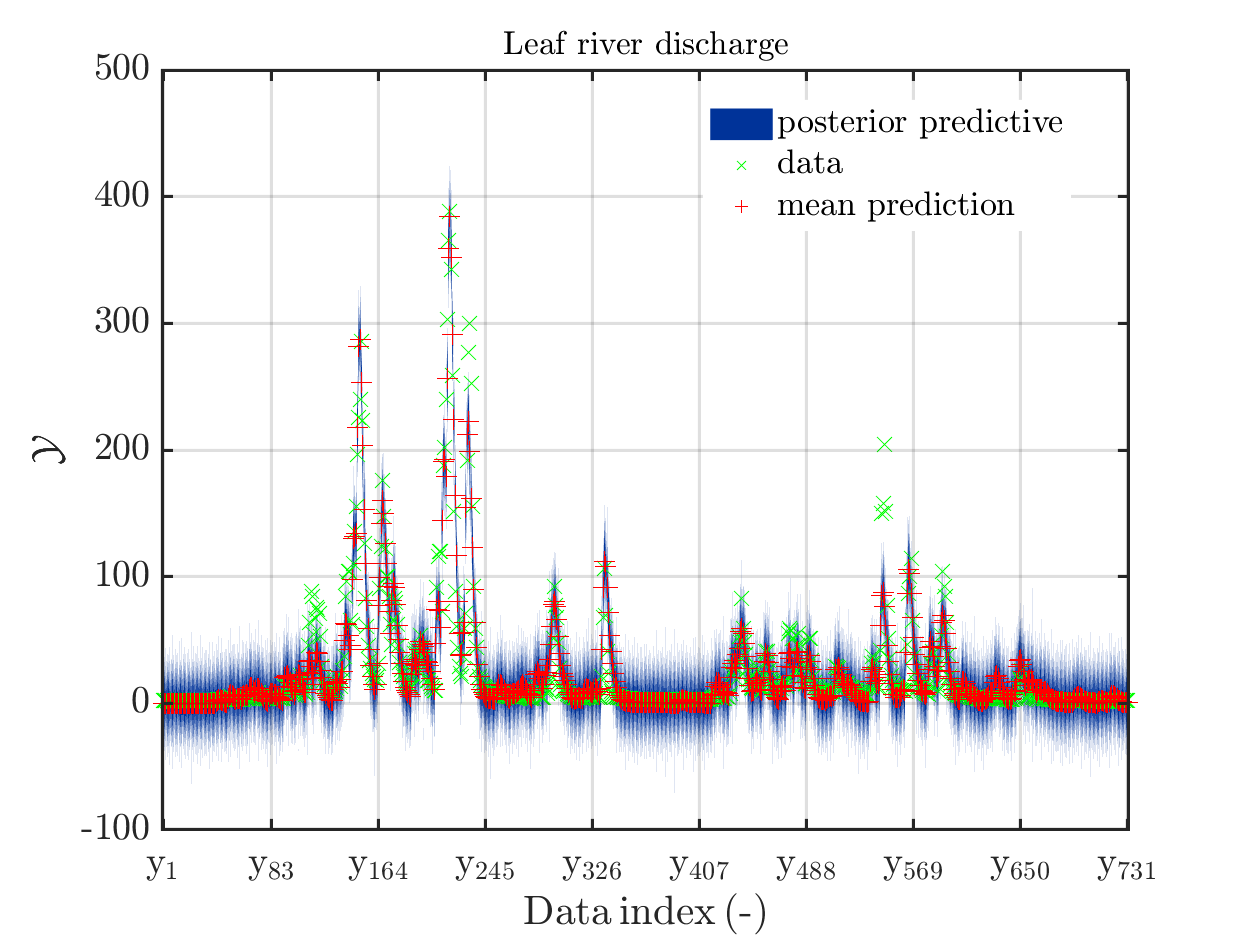
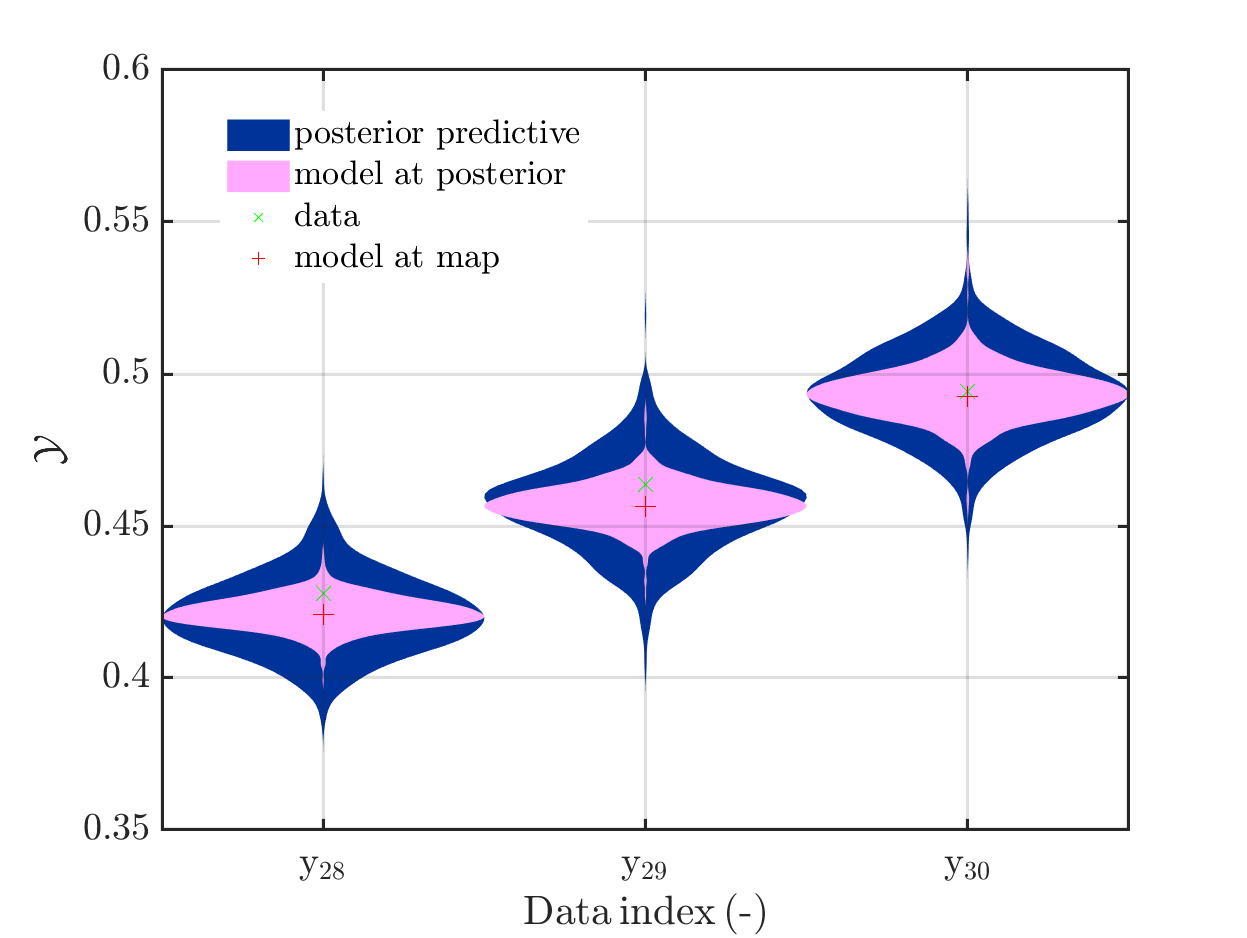
1. Legend “mean” / “mean prediction” in plots
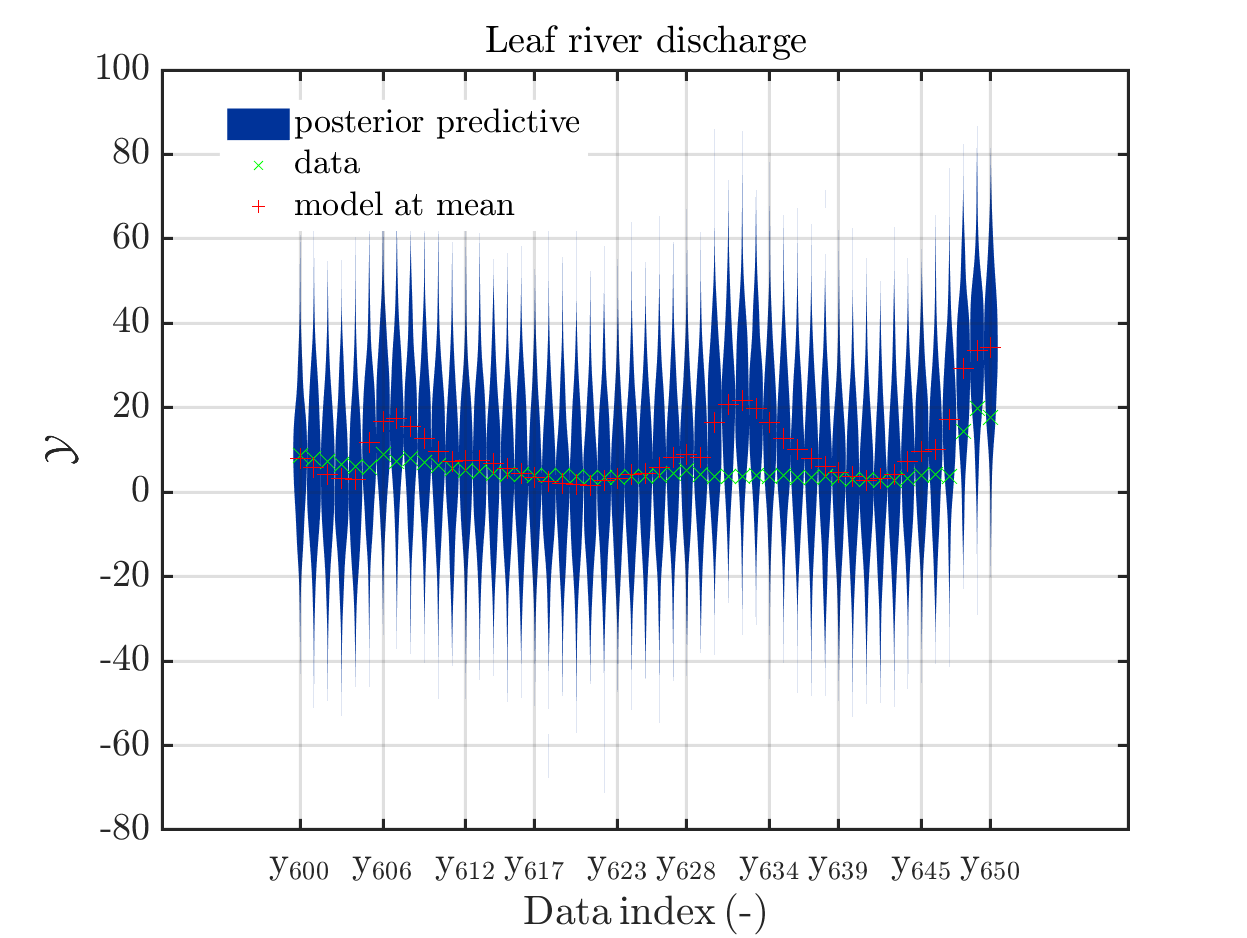
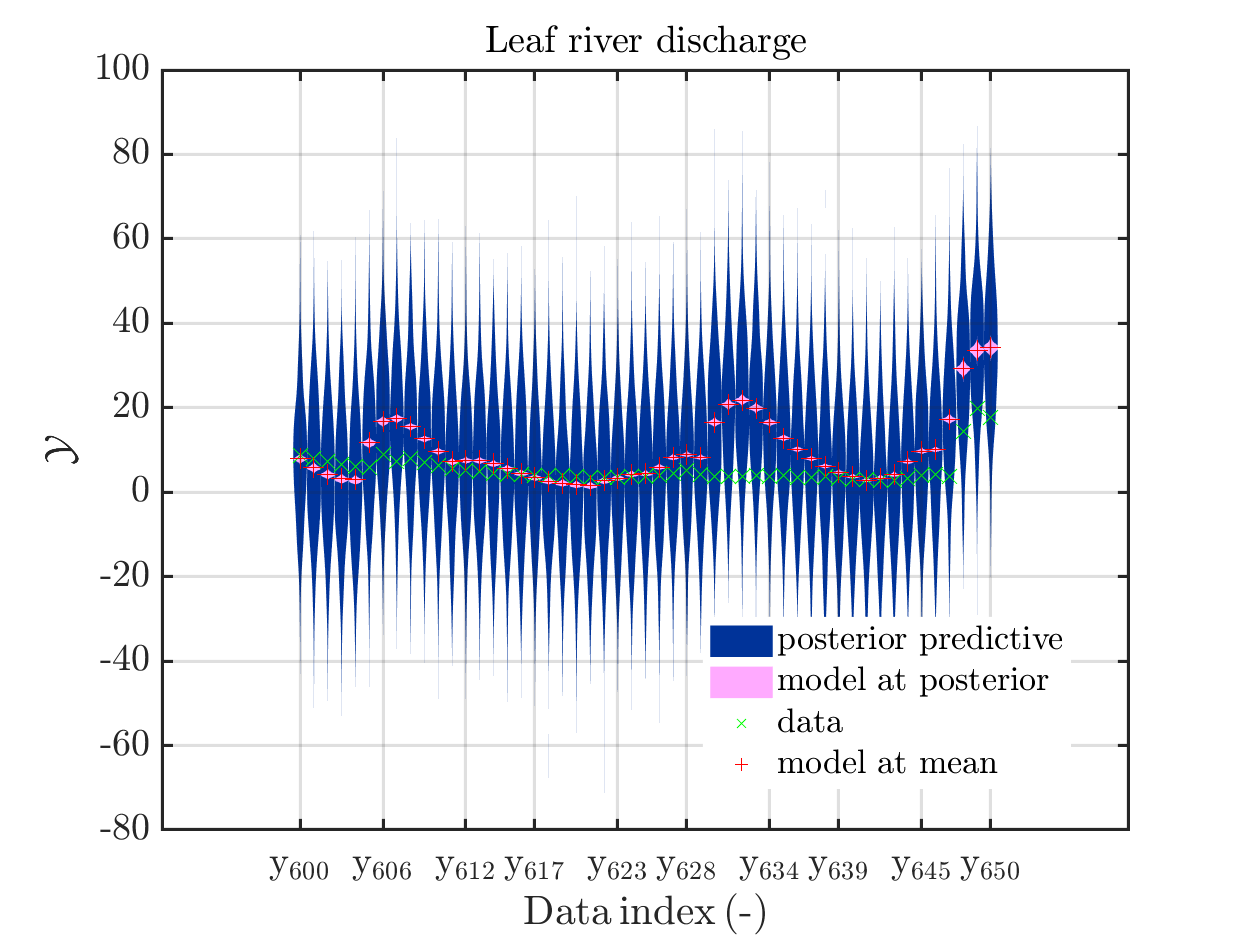
The current official version of uq_display_uq_inversion.m shows “mean” as legend for the red line in the plots for the predictive distribution to indicate
that the red line marks the value for the model output at the approximation for
\mathbb{E}(X|\mathcal{Y}) that is derived by computing the empirical mean of the samples for the model parameter generated from appropriate MCMC samples, at least according to my current understanding of the program code.
I must admit that I believed for some time that this “mean” is just the mean of the samples generating the shown predictive distribution.
In a preview version I got for uq_display_uq_inversion.m (thanks, Paul-Remo), the legend text “mean” is replaced by “mean prediction”, which is an improvement. But, I thins that also this notion could create confusion, since in many papers “mean prediction” corresponds to the mean of the derived predictions but here it is supposed to be the prediction derived by the evaluation the model at the mean of the samples for the model parameter generated from appropriate MCMC samples,
When I discussed results of my computations with a cooperation partner, being a UQ-expert, he asked me, which of these two interpretation of “mean prediction” is valid in my plots. Hence, it is proved that the notion “mean prediction” can create confusion and should therefore be replaced by a different notation, like e.g. “model(mean)” or “model@mean” or “model at mea” or …
2.) Inconsitent use of notion “point estimate”, fix of notations
I must admit that I am confused about the usage of “point estimate” in the manual,
in the program output and in the program code:
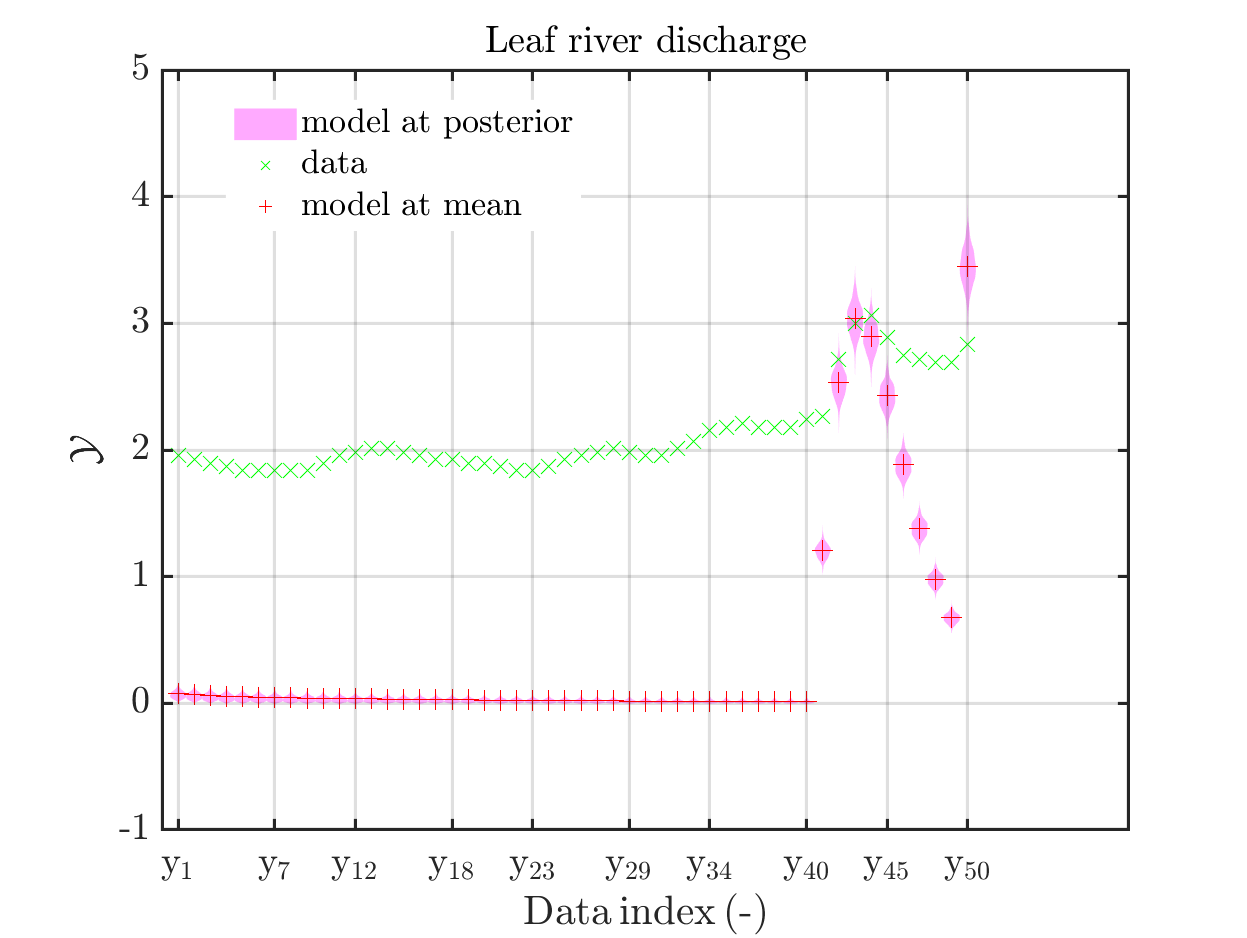
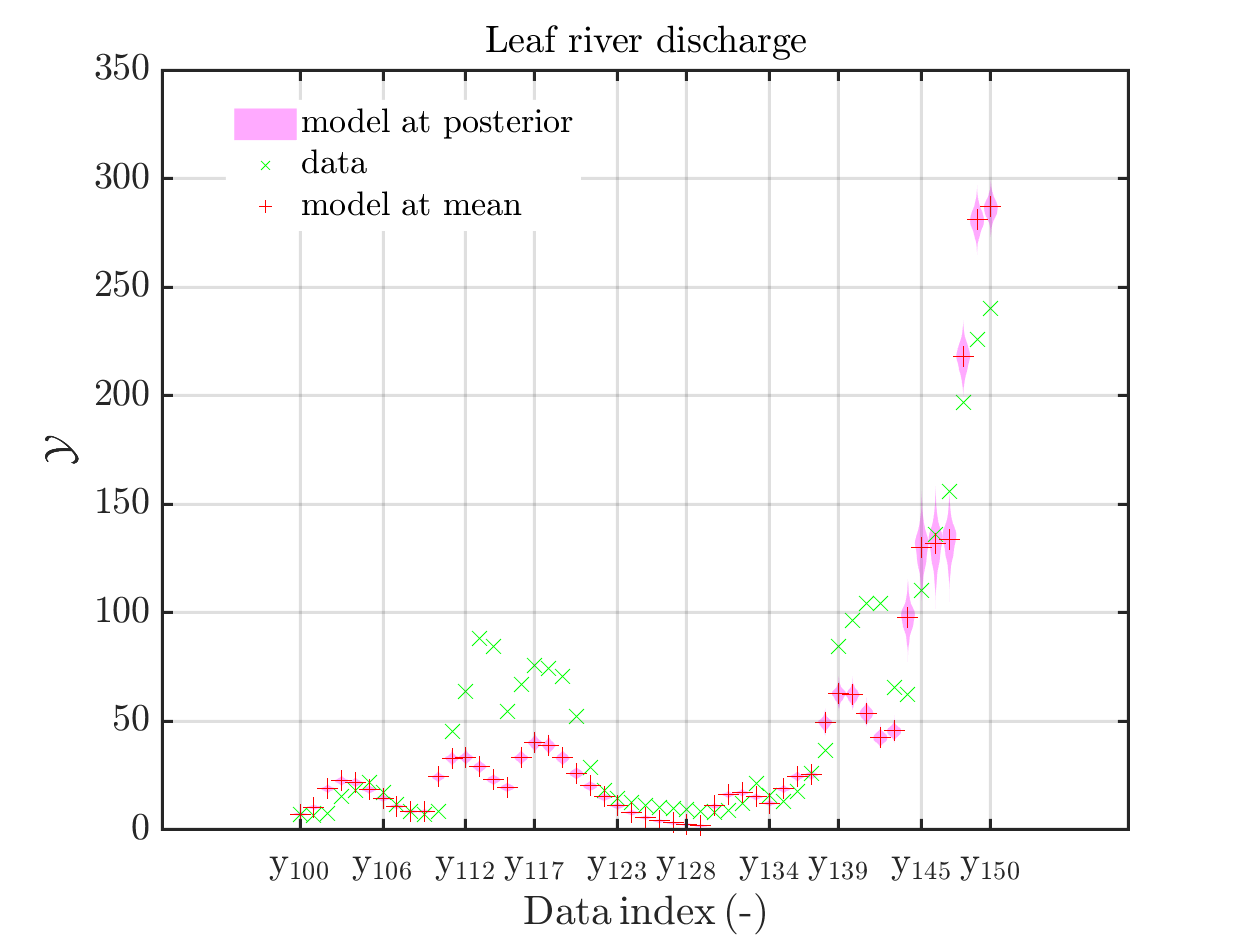
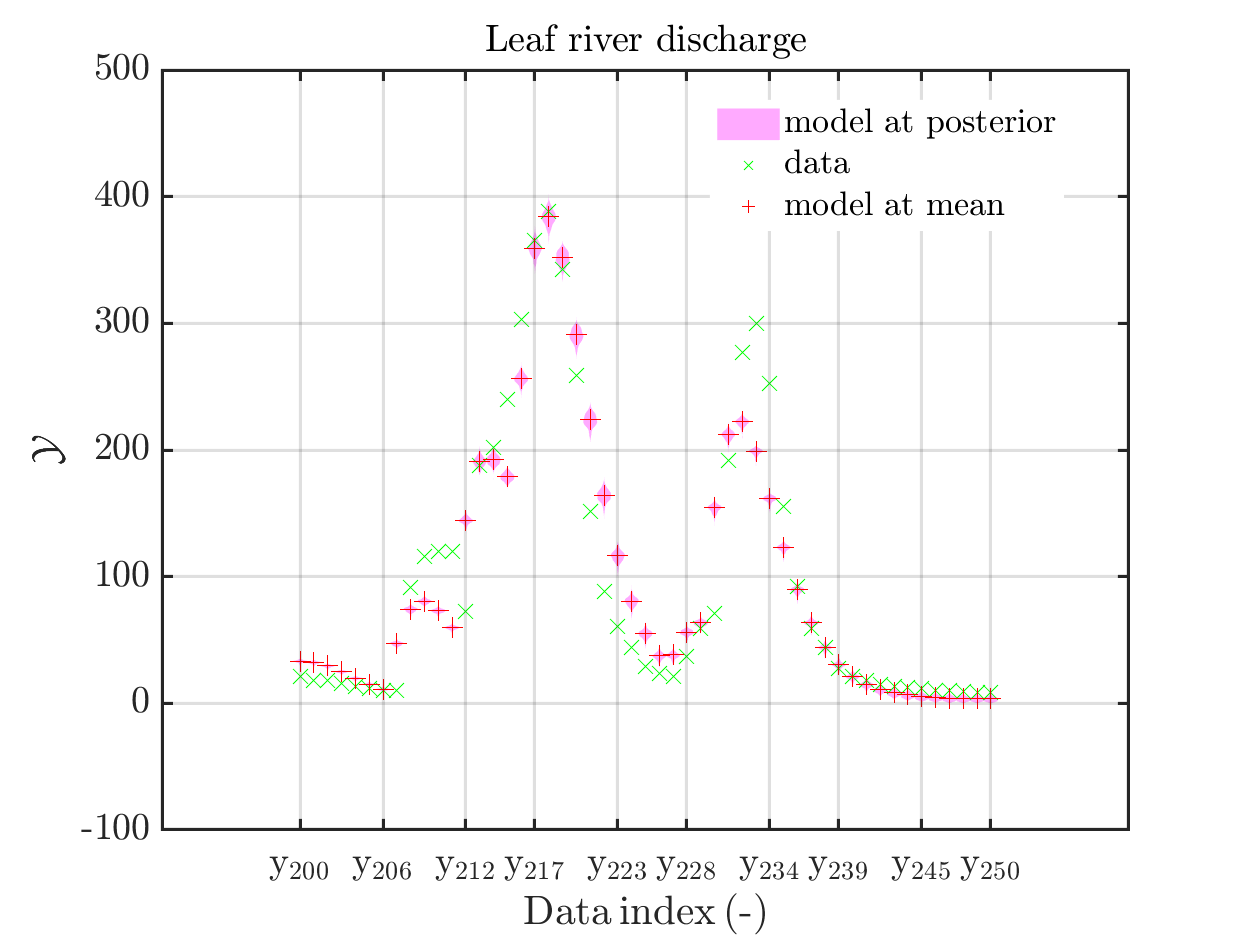
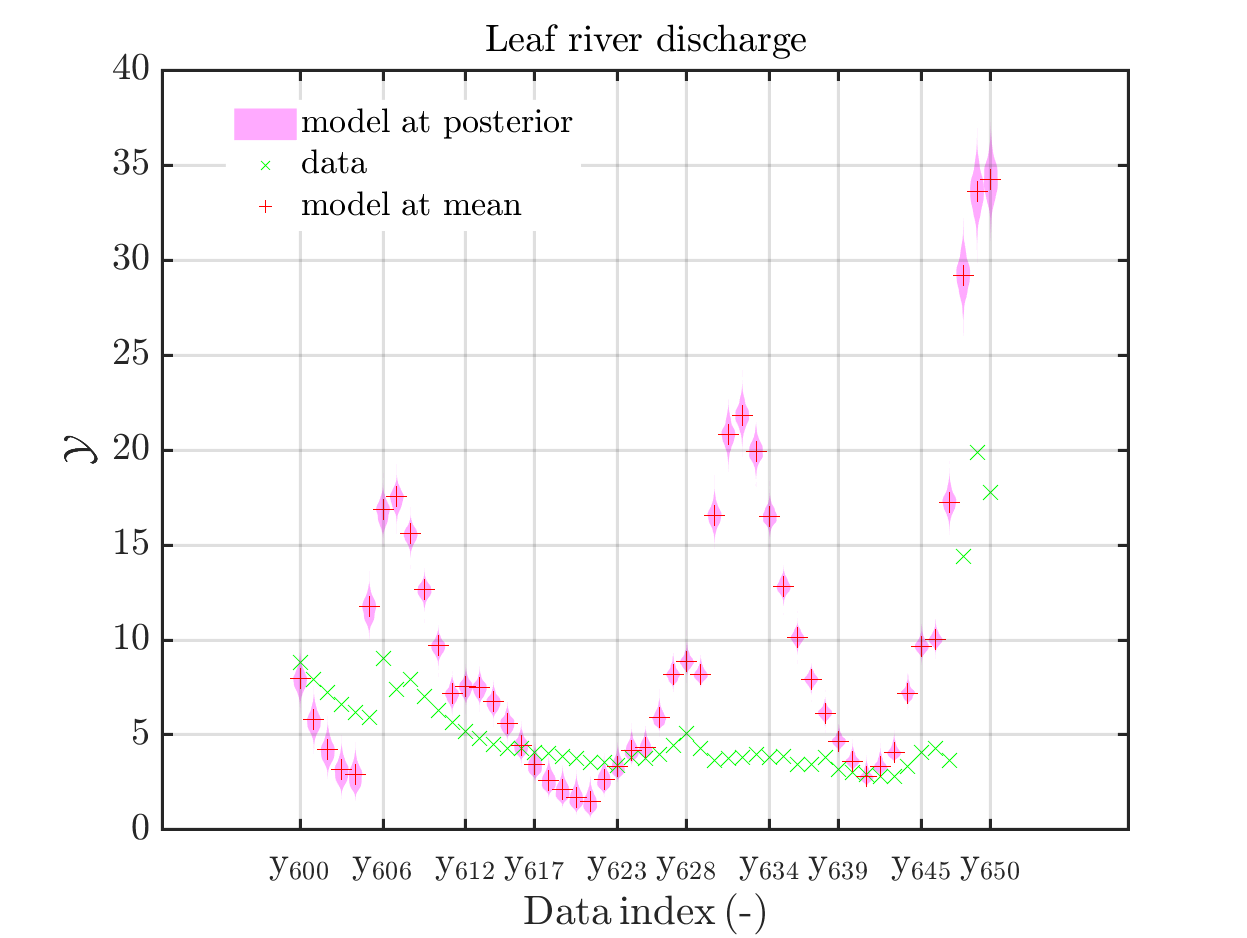
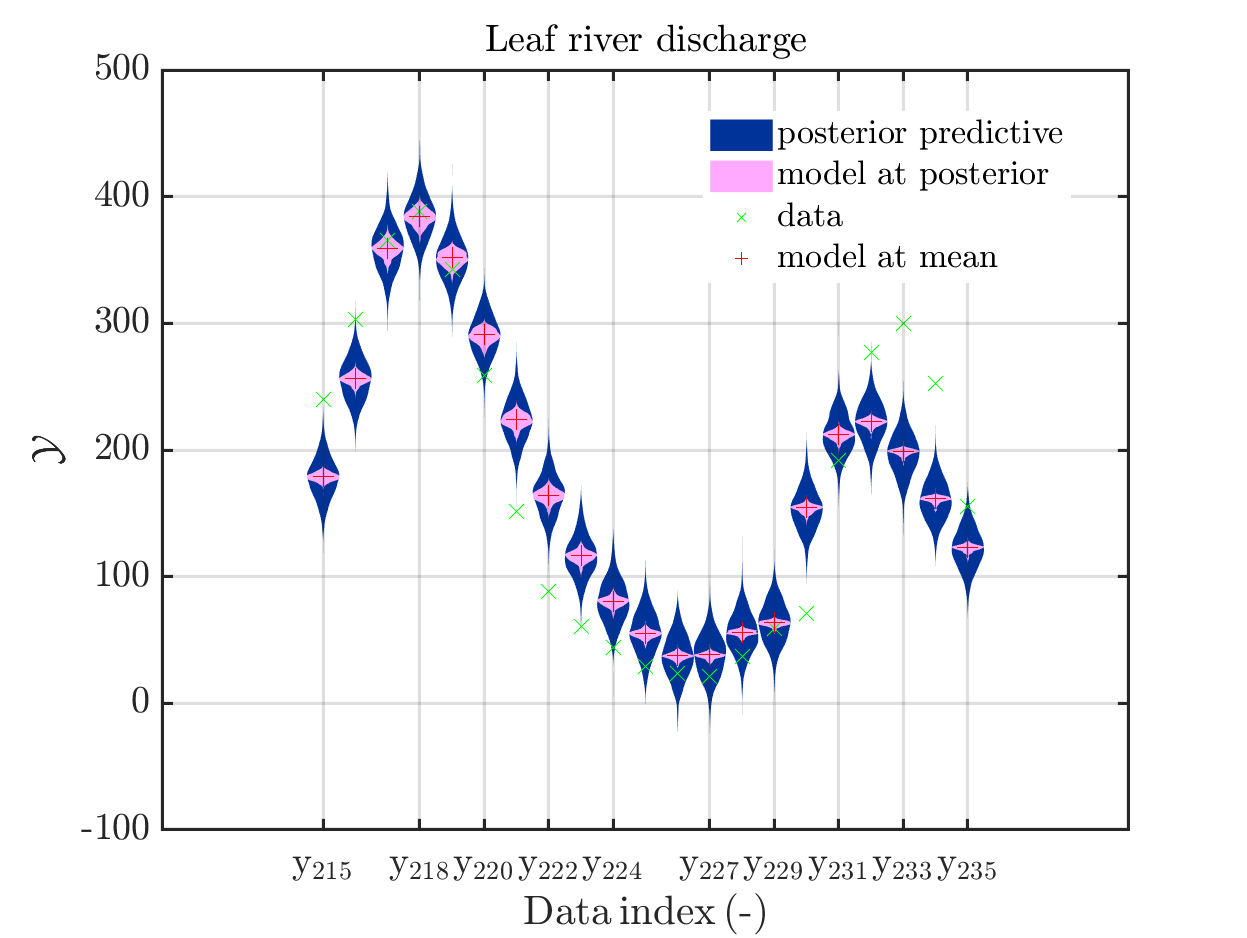
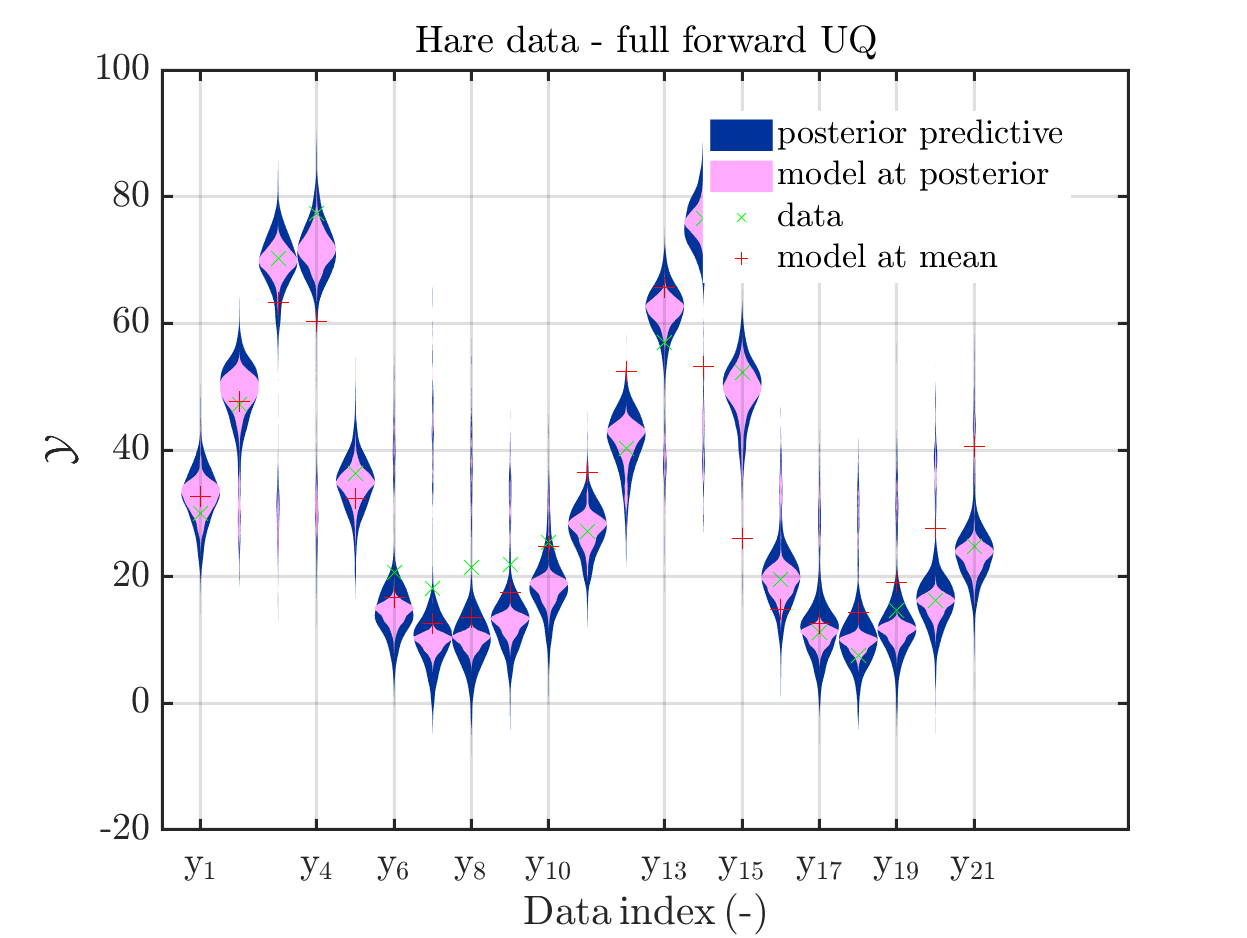
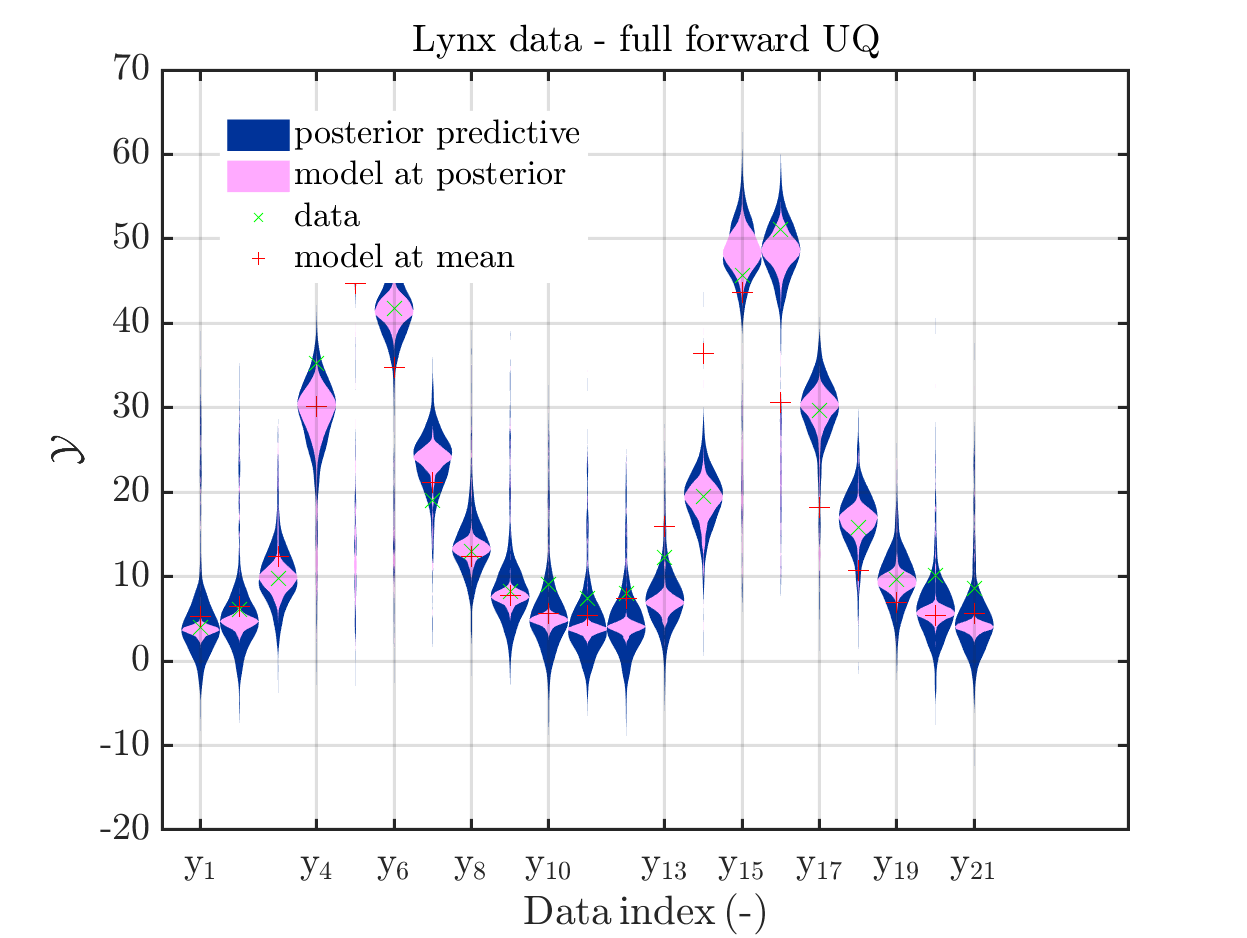
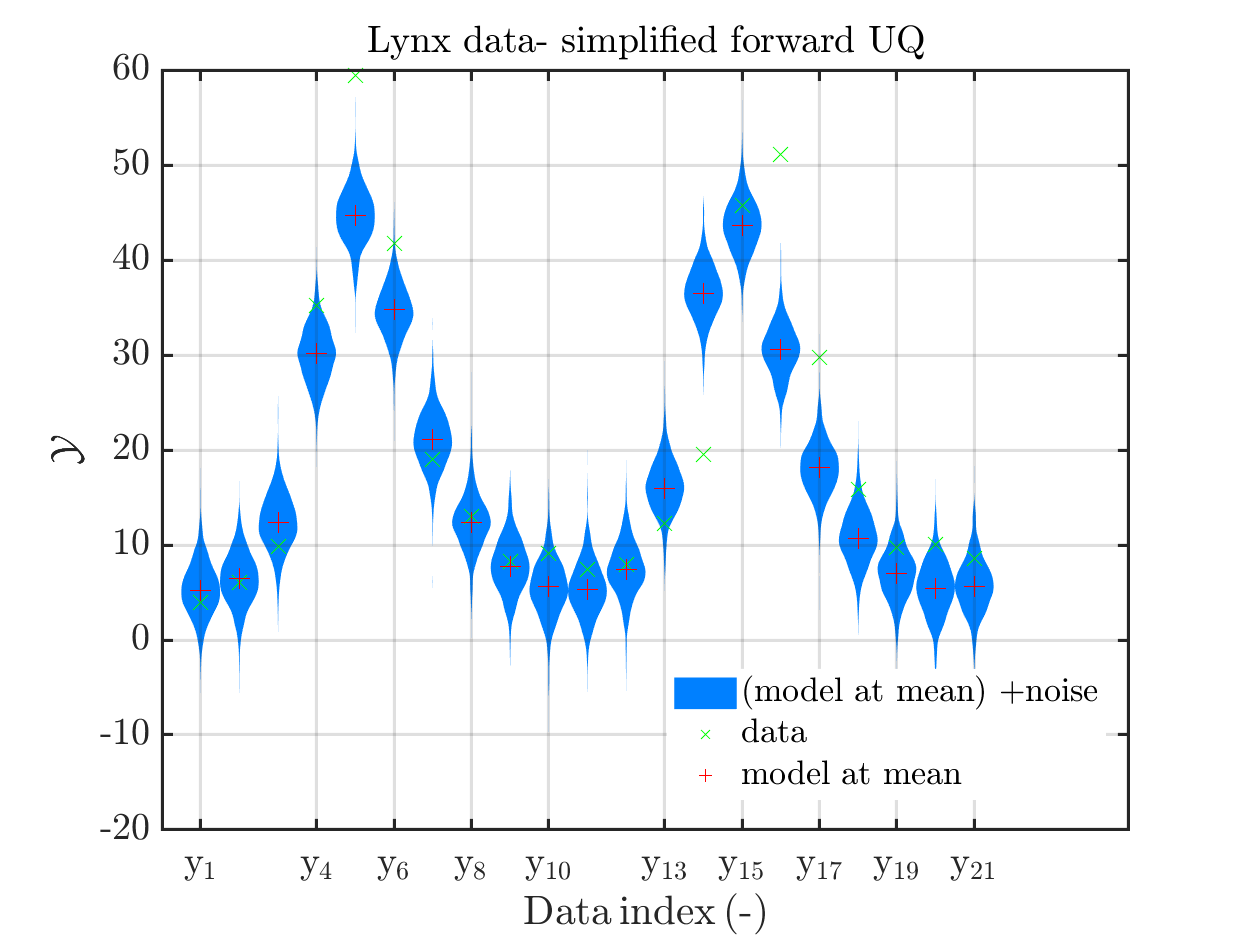
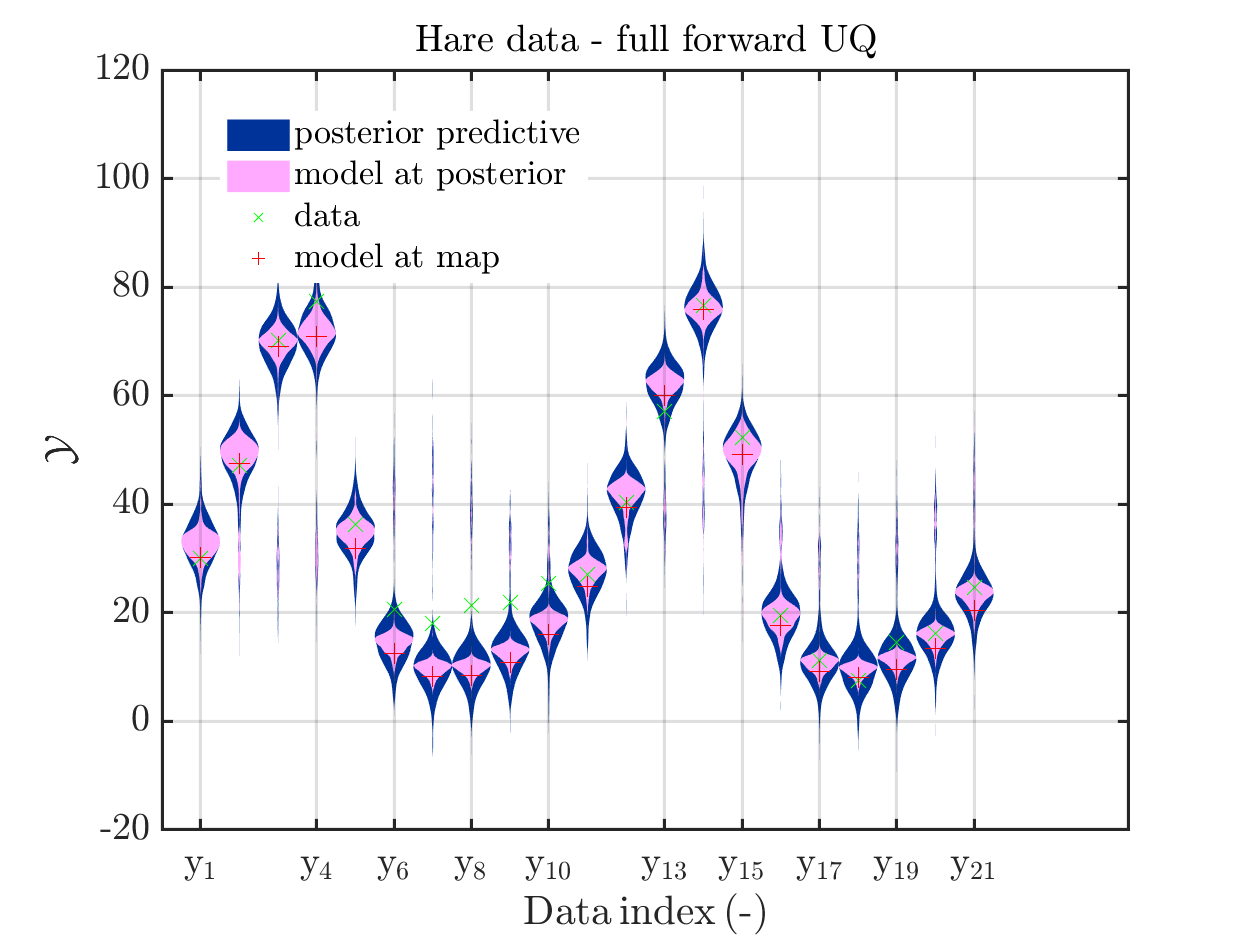
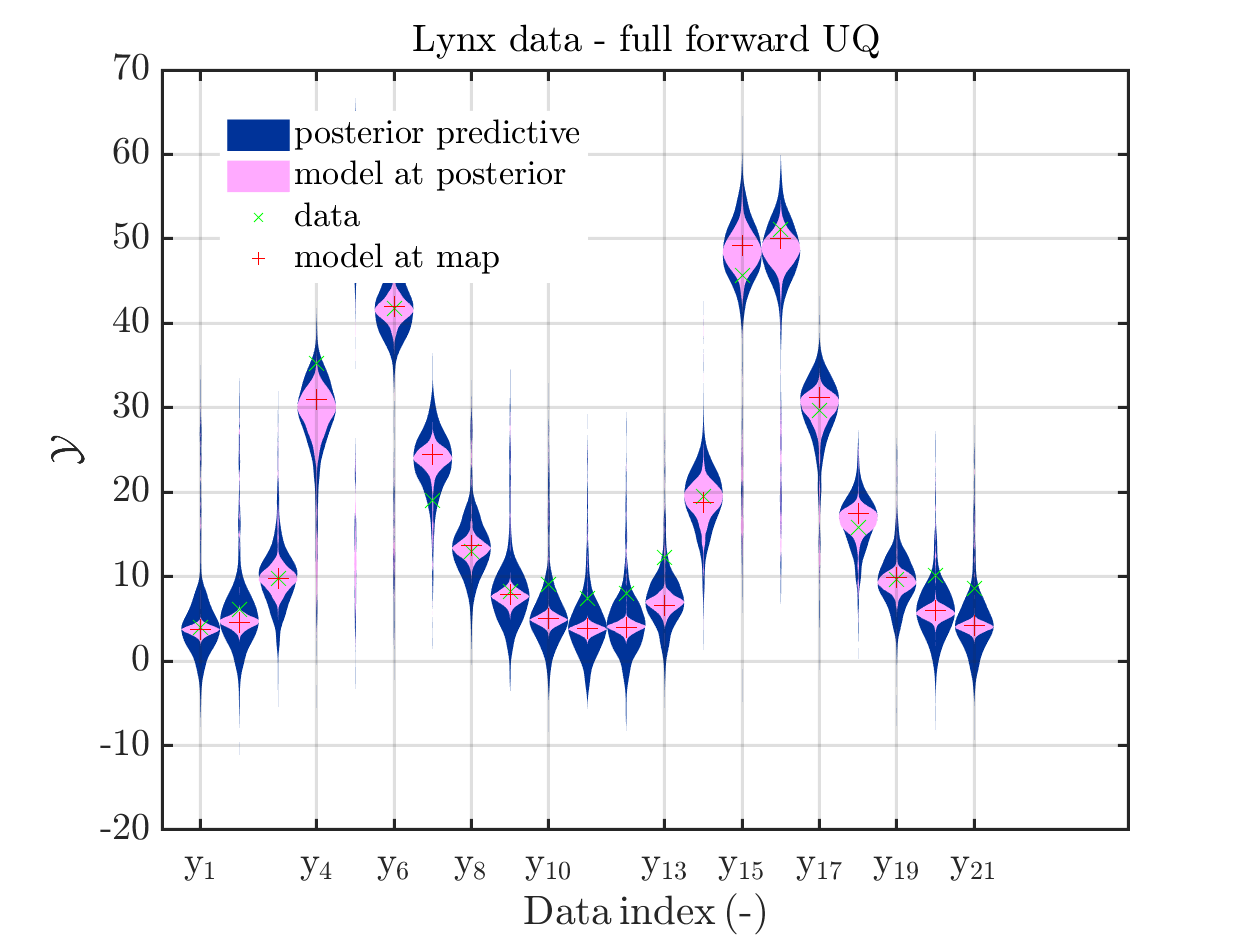
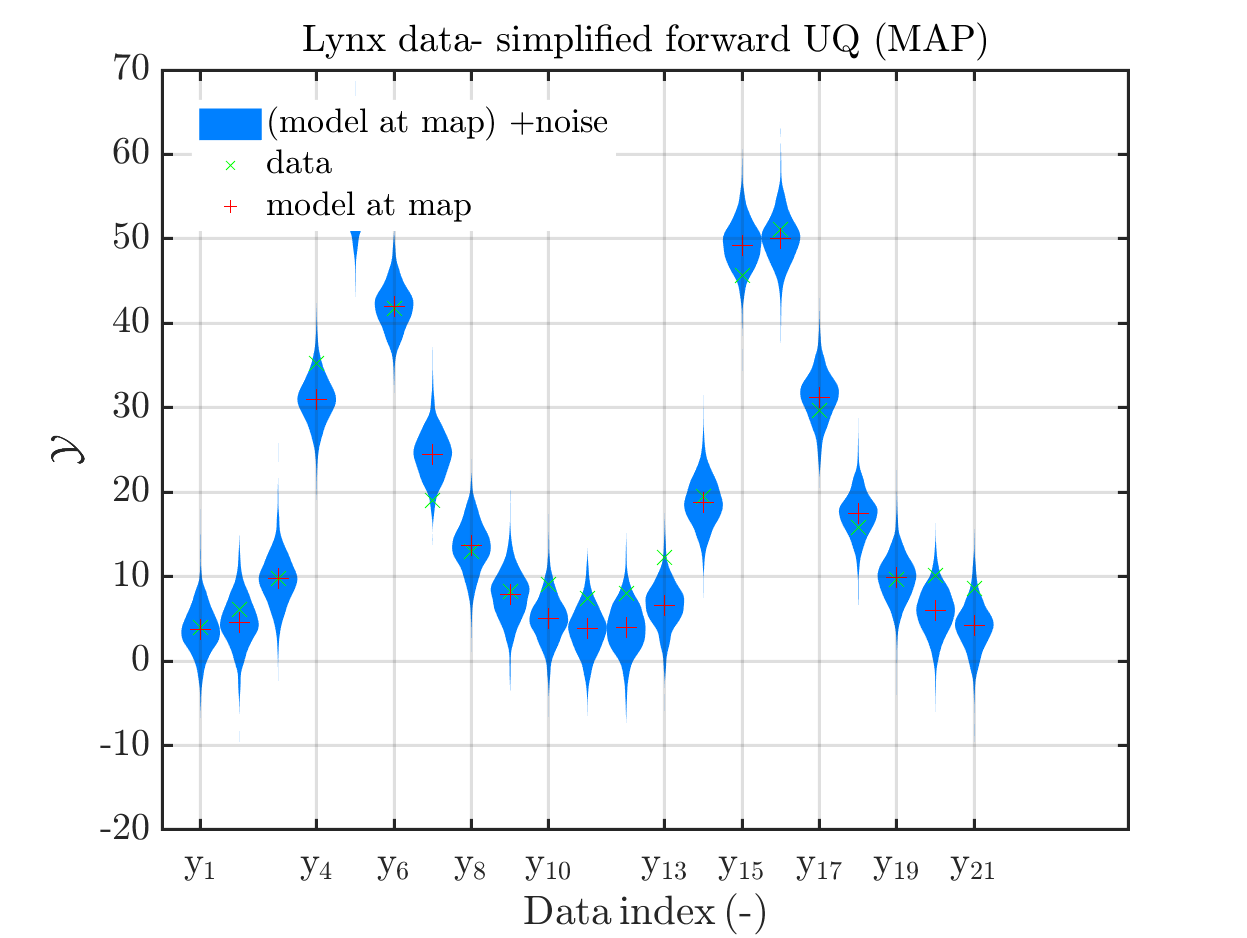
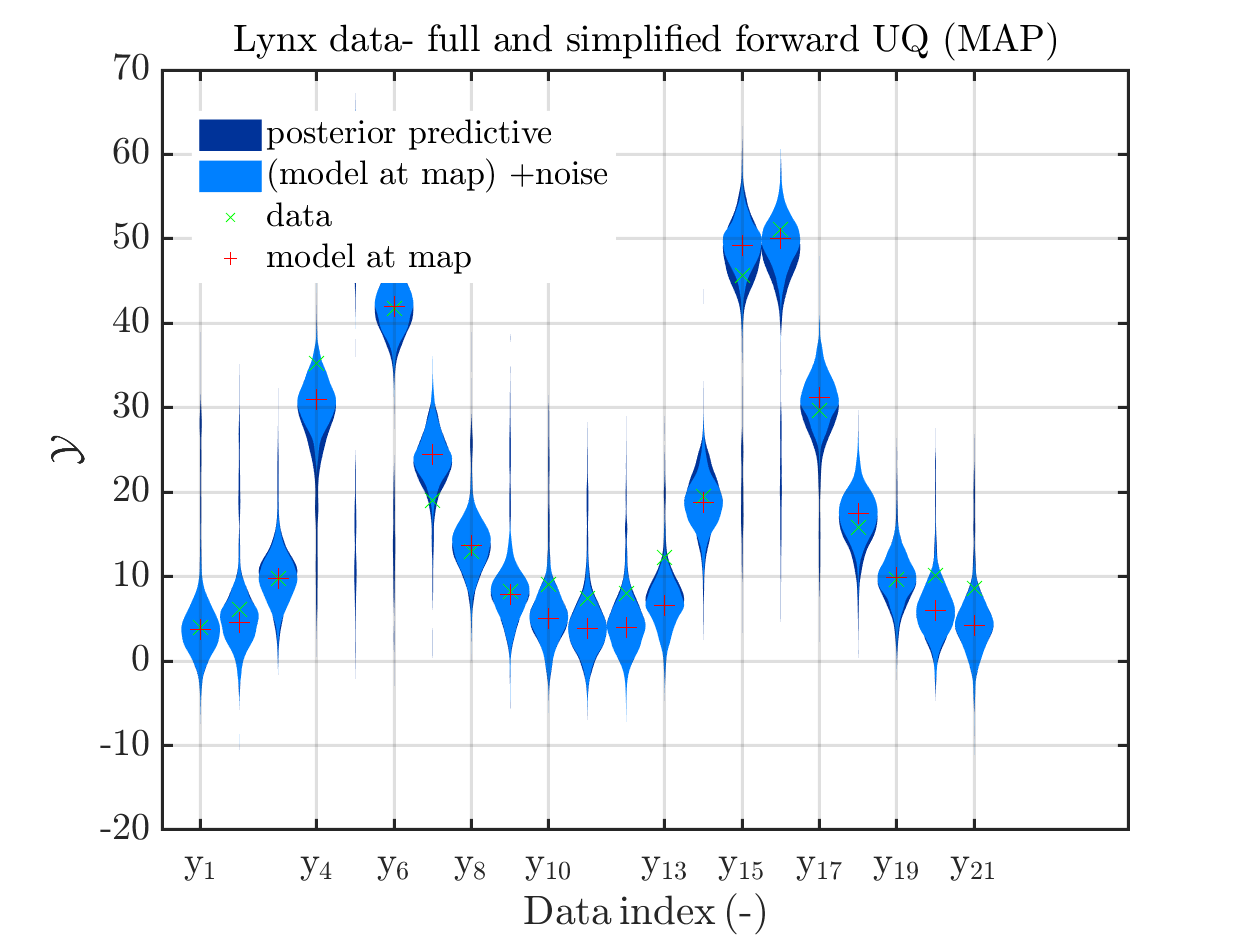
In view of the description of the plots in Fig 4(a), Fig 6(a), Fig 9(a) and the red mark in the plots, and what is shown in this plot according to the program code, “mean point estimate” (this notion is only used in these descriptions) seems to indicate the model output at the mean of the samples for the model parameter generated from appropriate MCMC samples.
But, in the description of Fig 4/6/9 it is also claimed that ``the empirical mean \mathbb{E} [X |\mathcal{Y}] estimated from the MCMC sample’’ is shown in Fig 4(a)/6(a)/9(a), which is not the case.
Hence, I wonder if maybe in both descriptions the mean of the samples for the model parameter generated from appropriate MCMC samples (that is shown Fig 4(b)/6(b)/9(b)) and the model output at this mean (that is shown Fig 4(a)/6(a)/9(a))
has been mixed up.
This would be compatible with an interpretation of " mean point estimate" in view of to the program output, other parts of the manual and the program code.
In the outputs of uq_print, see e.g. on the pages 21, 26, and 41, of the manual, it holds that there is a table entitled “Point estimate” and a row entitled “Mean”, providing the means of the samples for the model parameter and for the discrepancy generated from appropriate MCMC samples. Hence, it seems to be natural to denote this mean by “mean point estimate”.
Moreover, this interpretation seems to be compatible with the the discussion on emph{point estimator} on page 3 and the discussion near to equation (1.20).
Moreover, in view of the program code of uq_postProcessInversion, it holds that if the flag pointEstimate_flag is true, and the value of pointEstimate is “mean” the resulting value for module.Results.PostProc.PointEstimate.X is the mean of the samples.
I have not been able to find a general established interpretation for “mean point estimate” in my UQ books or by a Internet search .
I suggest to fix a notion for the mean of the samples for the model parameter
and for the model out[t at this mean in the UQLab User Manual for Bayesian inference and use it consistently.
To fix the notion, I suggest to add a new section 1.3.6:
To simplify my argumentation, I will use the notion therein in my text afterwards. following.
1.3.6: Quantities derived from results of a MCMC simulation
After performing a MCMC simulation, and a reduction of the derived sample due to
the burn-in and, if necessary, removing of bad chains (see Section 3.3), one can draw a set of samples from the remaining samples in the reaming MCMC chains. From these extracted samples one can compute a number of quantities.
In the following, we use the following notions to describe some them:
-
mean point estimate:
the empirical mean of the parameters values in the extracted samples, being an
approximation for the posterior mean, i.e. for the empirical parameter mean \mathbb{E}[X | \mathcal{Y}] as in (1.20) -
model output at mean point estimate:
output of the model if it is evaluated at the mean point estimate -
model(mean) :
legend text used in plots indicating the model output at the mean point estimate -
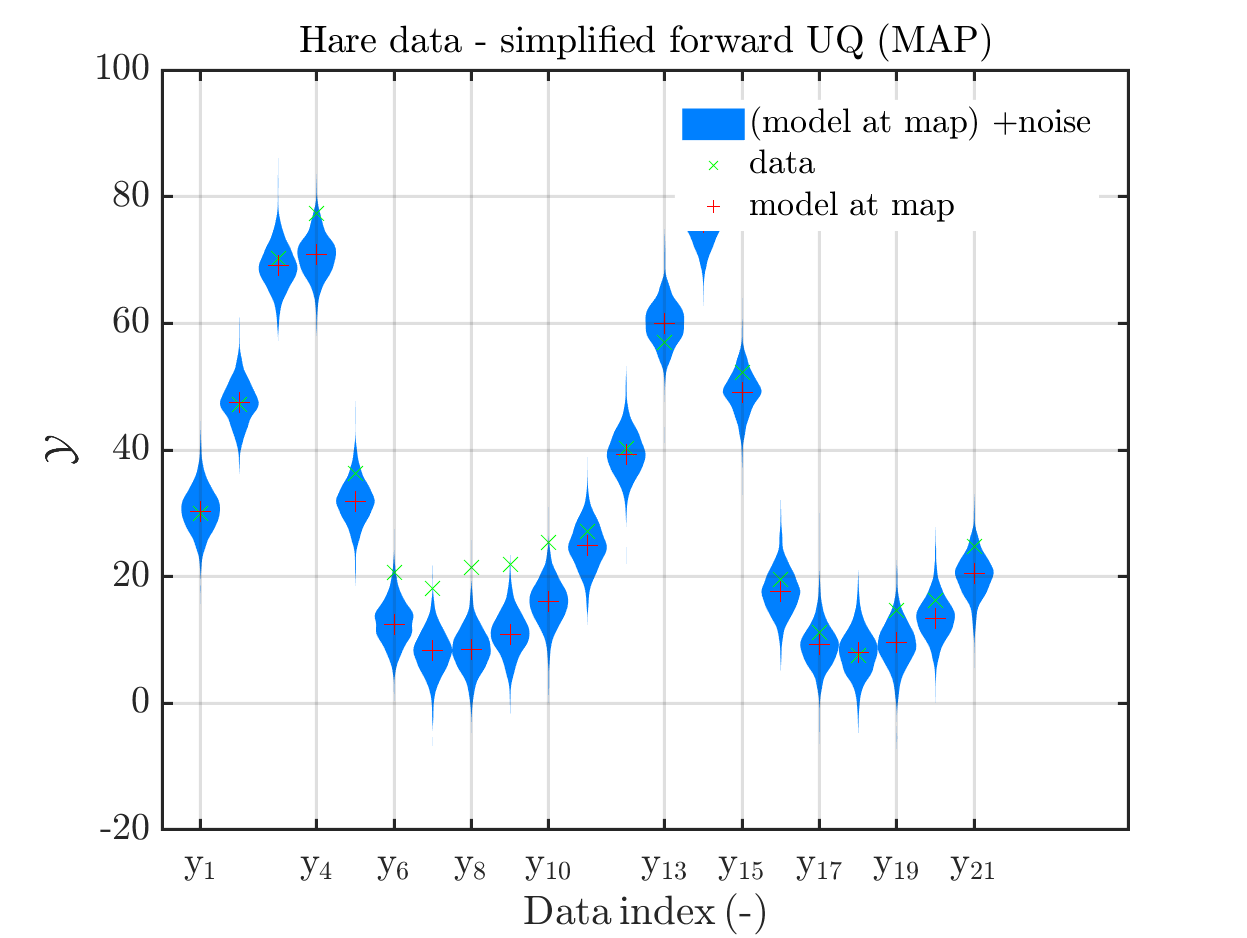
MAP point estimat:
set of parameters values in the extracted samples where the maximum of the posterior over all these samples is attained. This is an approximation of the
posterior mode, a.k.a. the maximum a posteriori (MAP), -
model output at MAP point estimate:
output of the model if it is evaluated at the MAP point estimate -
model(MAP):
legend text used in plots indicating the model output at the MAP point estimate
The samples for the posterior sample points are derived form the samples extracted from the MCMC samples following the considerations in the end of Sections 1.2.5 and 1.2.6. [My suggestion for a new section 1.2.6 follows below in point 7.]
3) Suggetion to add references to descriptions of plot contents
I suggest to add to the description of the plots in Fig. 4, 6, and 9, a reference/ references to the description(s) of the contents of the plot that can be found in Section 1 (e.g. to the new Section 1.3.6 if you decide to follow my suggestion).
This would help thereader to find these information especially if she/he needs information on the plot contents to understand some unexpected results in the plots in her/his application, and had forgotten during the programing period the details of the content of section 1.
4) Inconsistency between plot and its description
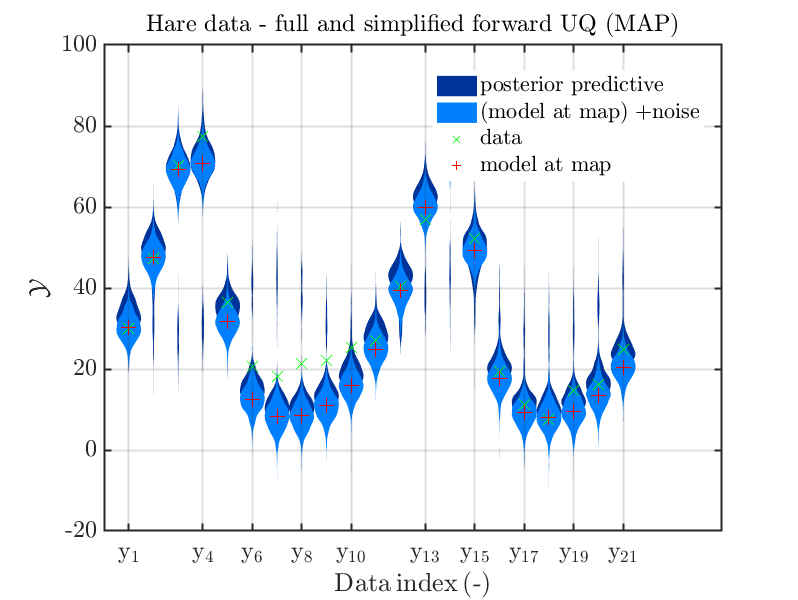
Page 23, Figure 4(a): In the current plot, there is no prior predictive distribution shown, but listed as content of the plot in its description.
Hence, I suggest to adapt either either the description or the plot.
5) Adding a legend to scatter plots
When dealing with scatter plots, it would be helpful if in
the plot there would be a legend indicating if
the mean point estimate or the MAP point estimate is marked.
6) Emergency meassures if plot legend hides important parts of plot
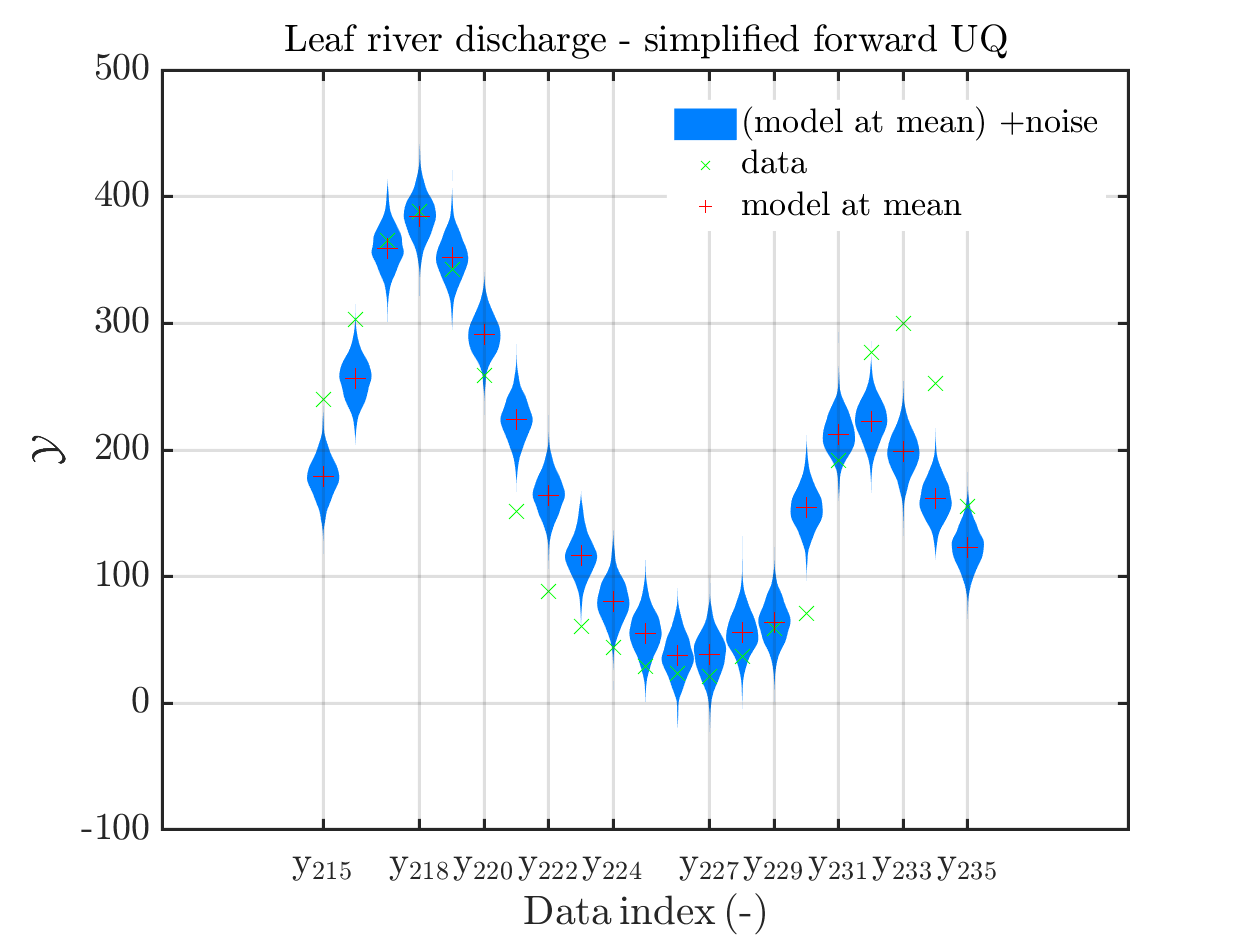
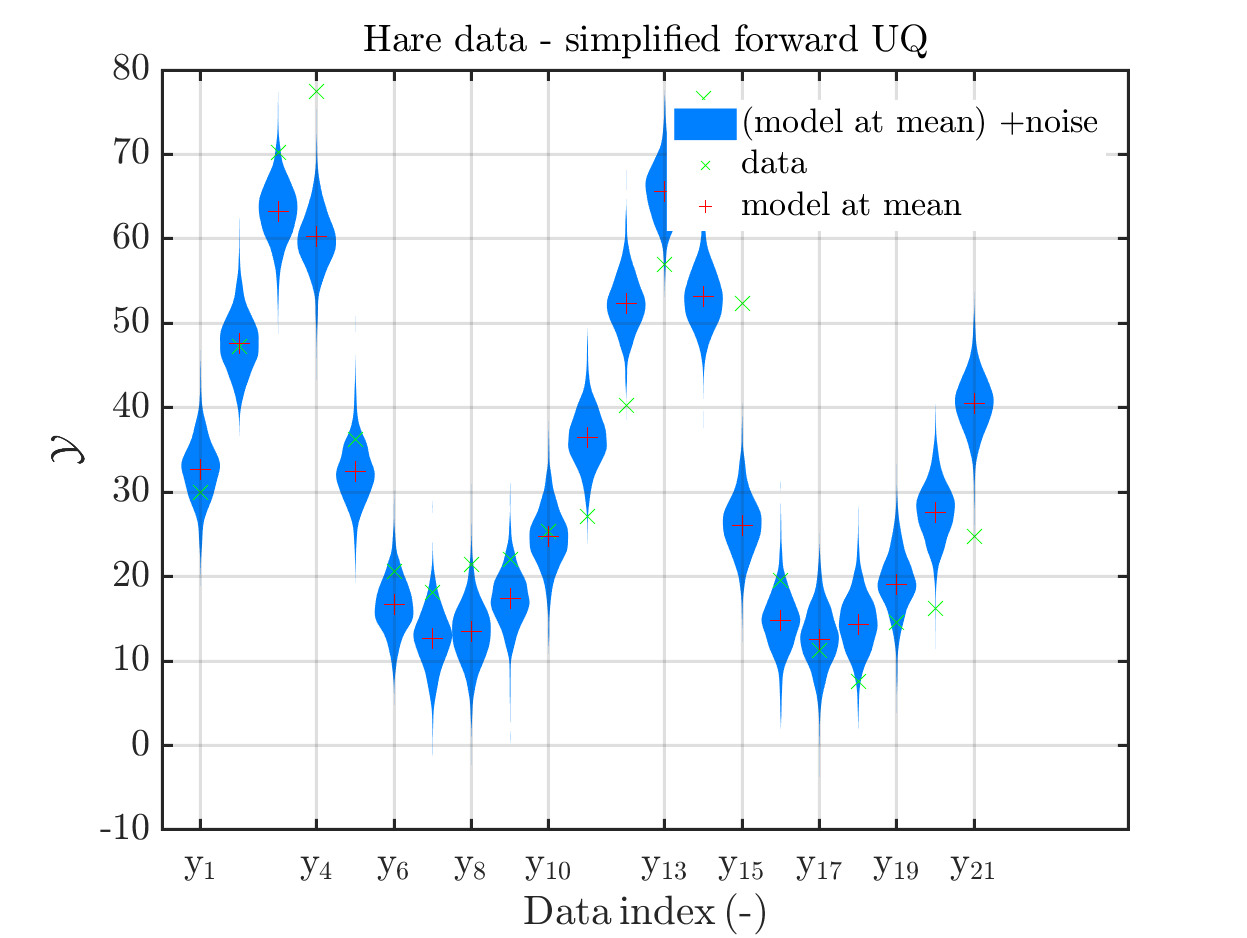
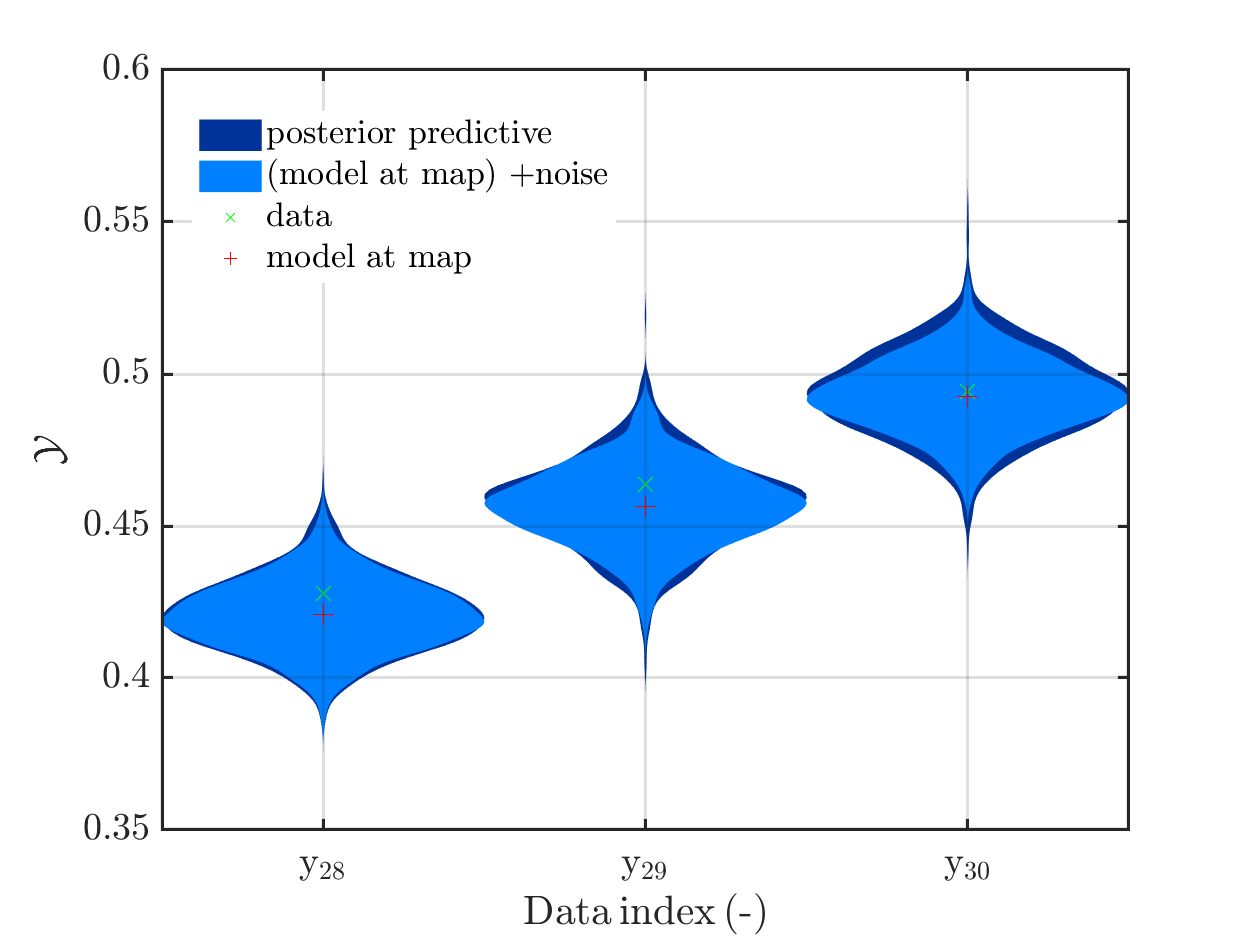
When I prepared my Case study Example of Bayesian inference computation creating a model output at the mean of the parameter samples that is outside of the discrete posterior predictive support I had to repeat the derivation of the example since in some cases the legend in the plot for the posterior predictive distribution plot was hiding the marks for the model output at the mean of the sample.
Since hiding of important part of the plots may also occur if one is dealing with the result of a fixed application, it may be helpful if there would be the emergency possibility to use a new option to the call of uq_print to either change the position of the legend within the plot or to show the legend in a separate figure.
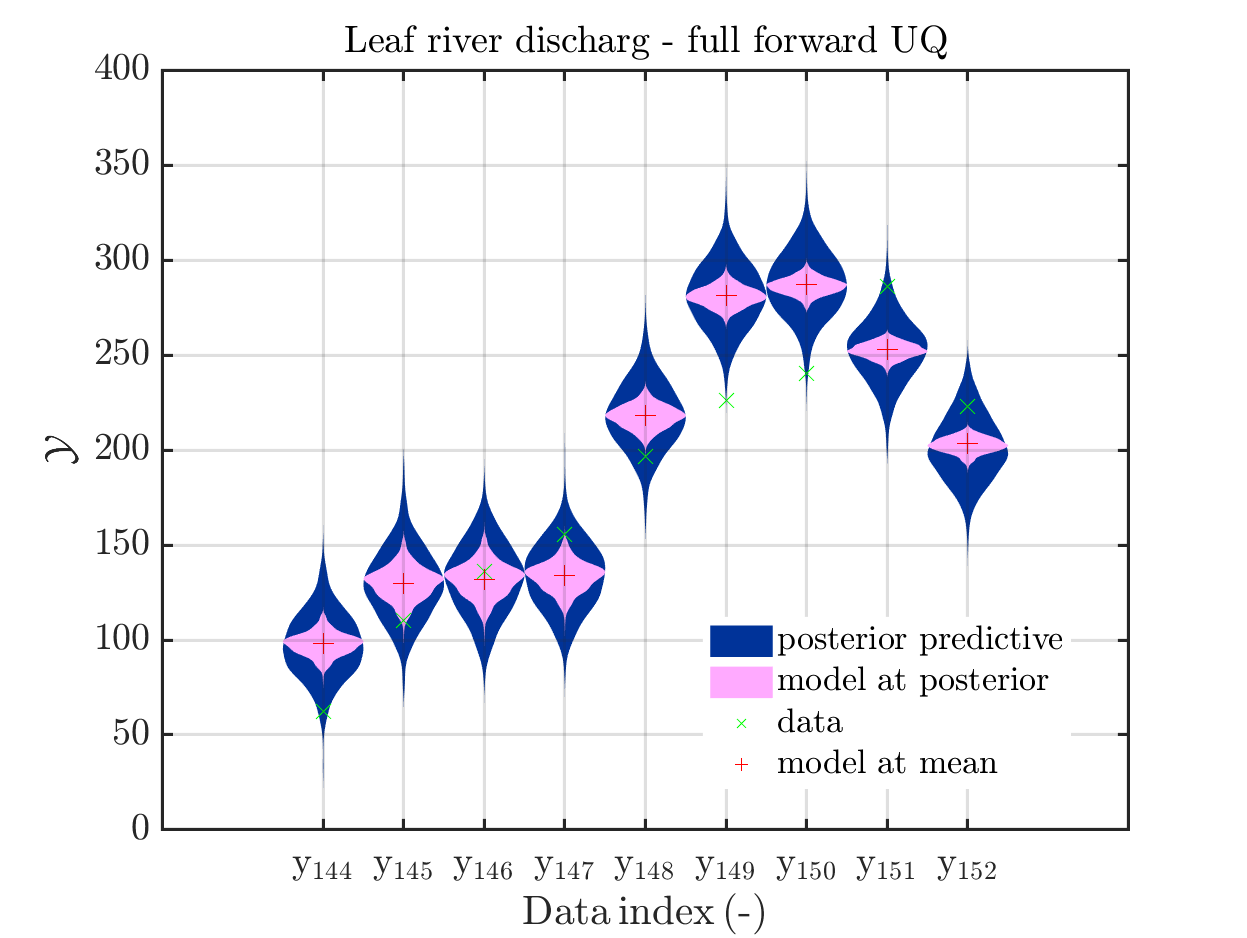
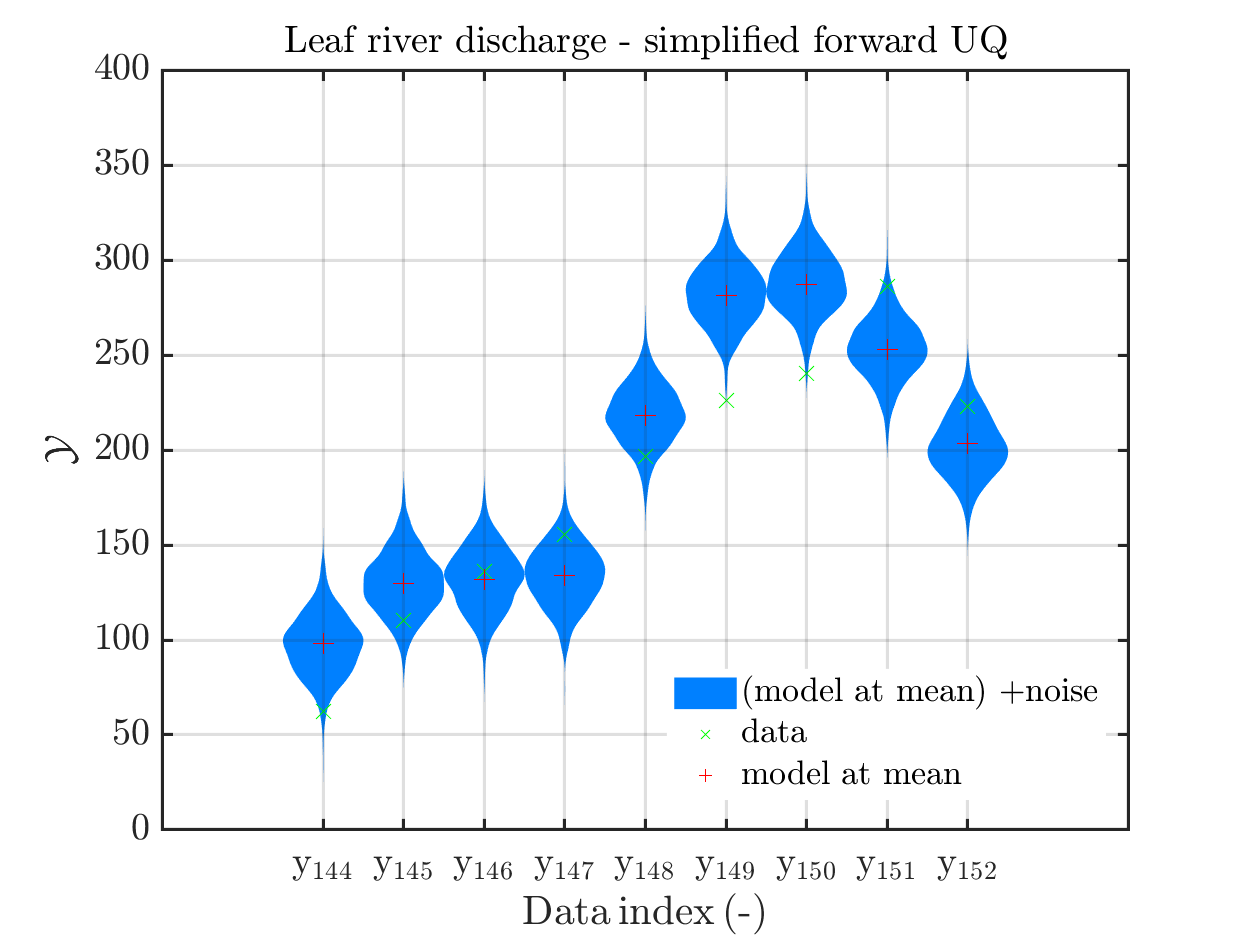
7) Formulation of Section 1.2.5 ``Model predeictions’’ only valid for a discrepancy with known parameters
I think that the current formulation of Section 1.2.5 ``Model predeictions’’ of
the manual is only valid for a discrepancy with known parameters, and does not
represent the calculations currently implemented in uq_postProcessInversion to derive a sample for the posterior predictive, if a discrepancy with unknown parameters is considered
For dealing with this situation, the following calculation is implemented:
Starting from samples x_i = \left( x_{i, \mathcal M}, x_{i,\varepsilon} \right) for \mathbf{x} = \left( \mathbf{x}_{\mathcal M}, \mathbf{x}_{\varepsilon} \right)
that were drawn according to \pi(\mathbf{x} | \mathcal Y), one evaluates \mathcal{M}(x_{i,\mathcal M}), and
draws independently samples from \mathcal{N}(\varepsilon|M(x_{i, \mathcal M}),x_{i,\varepsilon} ....) (I am not sure about the exact form that is possible for the matrix there) to derive samples for the posterior predictive.)
I suggest to change the title of of Section 1.2.5 to “Model predictions for discrepancies with known parameters” and to add a new section 1.2.6: *“Model predictions for discrepancies with unknown parameters”’’ dealing with this situation.
I hope that you like at least some of my suggestions.
Best regards
Olaf