I have run into a (probably ignorant) issue defining a new input when using the statistical inference module.
I have some data that looks to be lognormal and I am hoping to propregate the distribution for further bayesian inference.
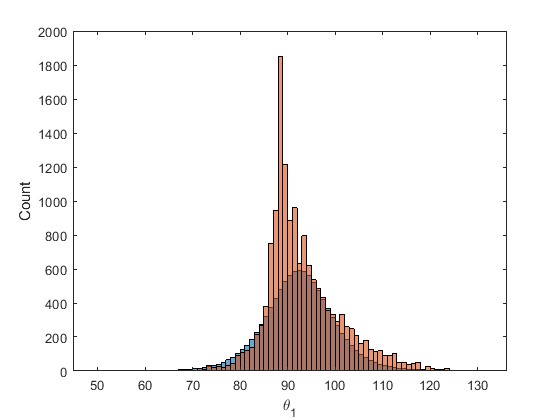
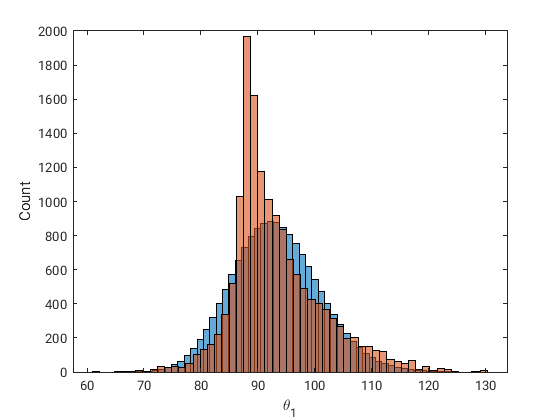
The red histogram is the output of the Bayesian inference and the blue is the infered marginal.
The relevant snippet of code is provided below. The data is uploaded as a zip file.
Any help would be greatly appreciated ![]()
I would like to add that I have tried all the inferences criterion, and defining various marginal families to no greater success than the default.
Regards,
Truong
UQLAB_QUERY2.zip (1.5 MB)
% Inference of the samples created in MCMC
PostSample3D = myBayesianAnalysis_surrogateModel.Results.PostProc.PostSample ;
PostSample2D = reshape(permute(PostSample3D, [2 1 3]), size(PostSample3D, 2), [])';
iOpts.Inference.Data = PostSample2D(:,2) ;
iOpts.Marginals.Type = 'auto';
iOpts.Copula.Type = 'Independent';
iOpts.Inference.Criterion = 'KS';
mydist_infer = uq_createInput(iOpts);
uq_print(mydist_infer)
%Creation of samples from the two post-inputs
Xtest_inf= uq_getSample(mydist_infer , 10000, 'LHS');
% % hold on to the existing plot and add a histogram of the second dataset
figure
histogram(Xtest_inf,'BinMethod','auto')
hold on
histogram(PostSample2D(:,2))
xlabel('\theta_1')
ylabel('Count')