Hello,
I have set a Bayesian inversion, with a multiple output model, but the posterior distributions of my inputs raise questions. I am not sure of the relevance of my results.
Here is a description of my analysis:
- I have two inputs, E and c, which follow lognormal distributions.
- My model comes from a finite element software (ZSoil), with two outputs. It is surrogated with a PCK metamodel (Eloo ~ 10^-6 for both outputs).
I use the following code to create my inputs :
InputDist.Marginals(1).Name = ‘E’;
InputDist.Marginals(1).Type = ‘Lognormal’;
InputDist.Marginals(1).Moments = [8000 2000];
InputDist.Marginals(2).Name = ‘c’;
InputDist.Marginals(2).Type = ‘Lognormal’;
InputDist.Marginals(2).Moments = [10 ; 2];
myPriorDist = uq_createInput(InputDist);
To perform the inversion :
DiscrepancyOpts.Type = ‘Gaussian’;
DiscrepancyOpts.Parameters = 0.5 ;myData.y = Observation (1,:);
Bayes.Type = ‘Inversion’;
Bayes.Data = myData;
Bayes.Prior = myPriorDist;
Bayes.ForwardModel = my_PCK;
Bayes.Discrepancy = DiscrepancyOpts ;
myBayes_stg3 = uq_createAnalysis(Bayes);
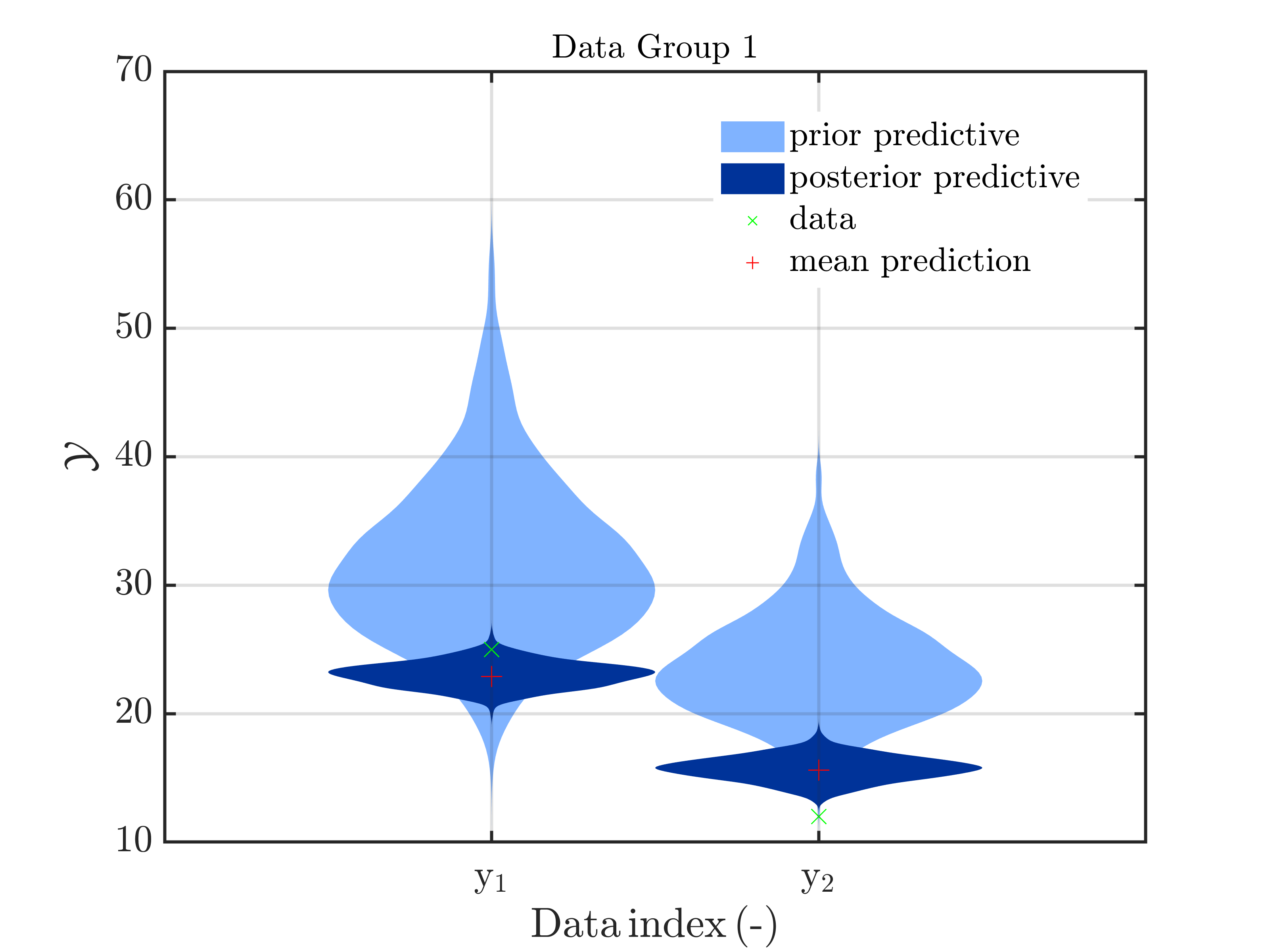
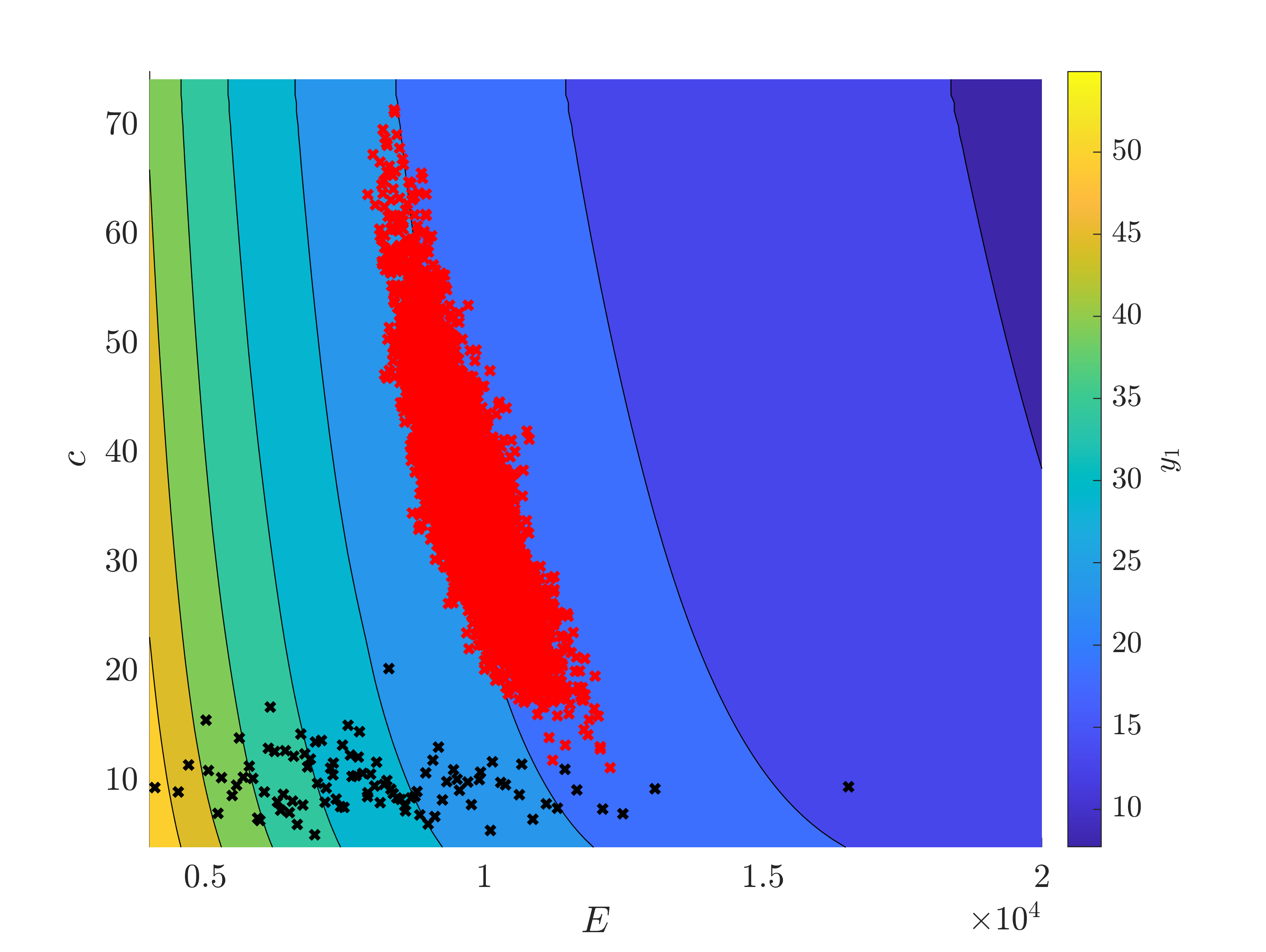
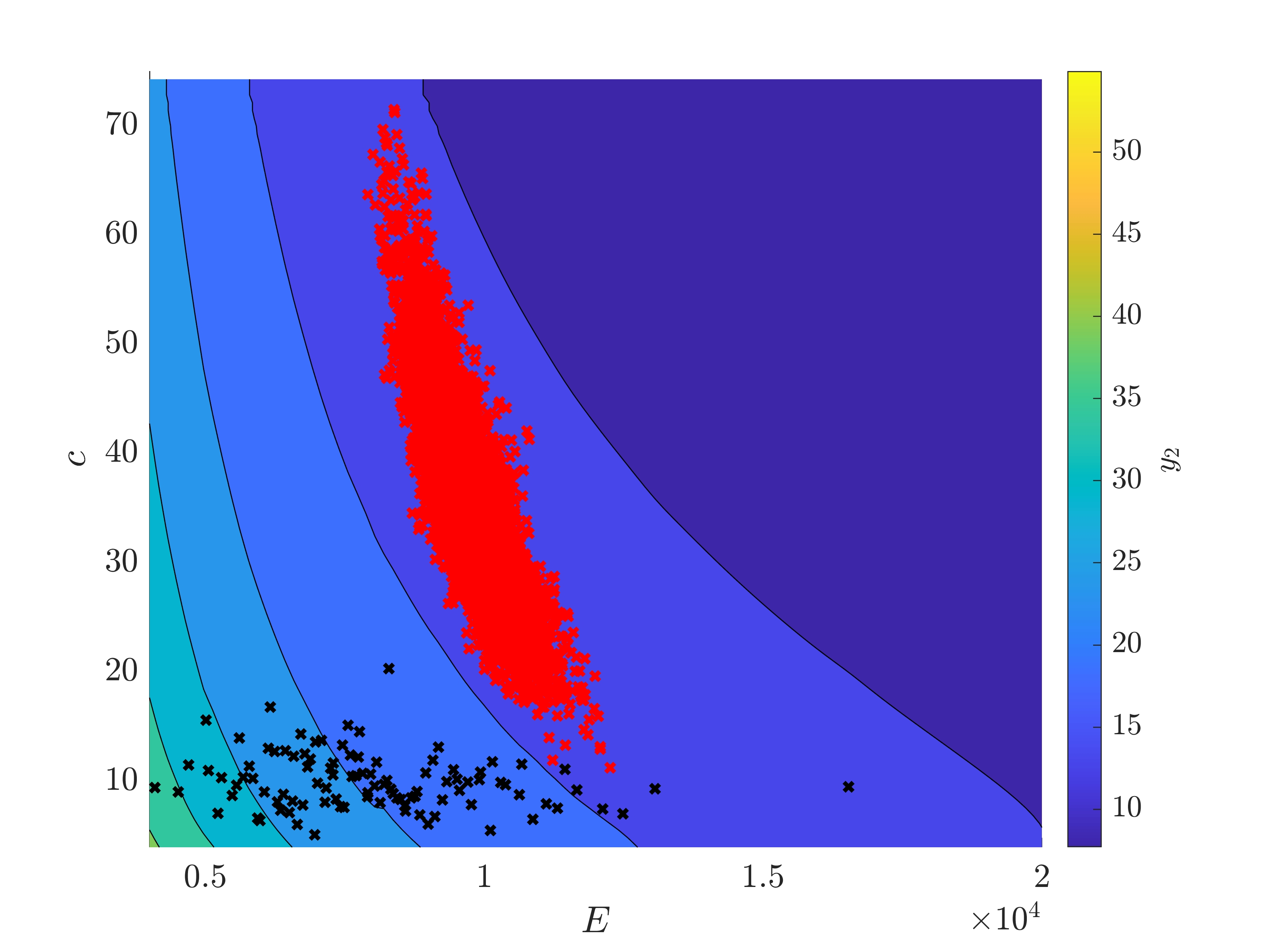
The measurement (25 ; 12) is quite far from the mean predicted value (30,22).
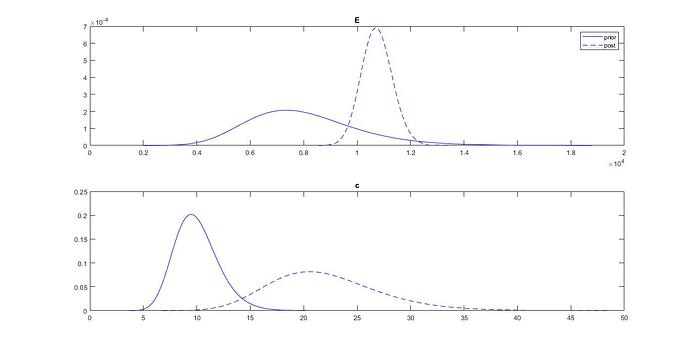
When I look at the prior and posterior distributions of my inputs, I have the following graph:
My question is : is it possible, after a Bayesian inversion, that the standard deviation of an input increases ? On the graph, we can see that the standard deviation of c is higher after the inference. I thought, as the measurement brings information, that the standard deviation should be reduced.
I also have a technical remark : when I set the distribution of c to “constant”, in order to study only the influence of E on the inference, the Bayesian inversion should run without updating c, as shows the user manual. It seems in this case, the following error appears (before the beginning of AIES’s algorithm) :
Unrecognized property ‘Model’ for class ‘uq_model’.
Error in uq_initialize_uq_inversion (line 394)
ForwardModel(ii).Model = uq_createModel(ModelOpt,‘-private’);Error in bayes_biais_uqlab (line 38)
myBayes_stg3 = uq_createAnalysis(Bayes_stage3);
It seems this message also appeared to @olaf.klein : is there something I can do to set c constant, without changing my model ?
Thanks a lot,
Marc Groslambert