Hello!
I conduce metamodeling using data driven based polynomial chaos expansion. I have some question about PCE.
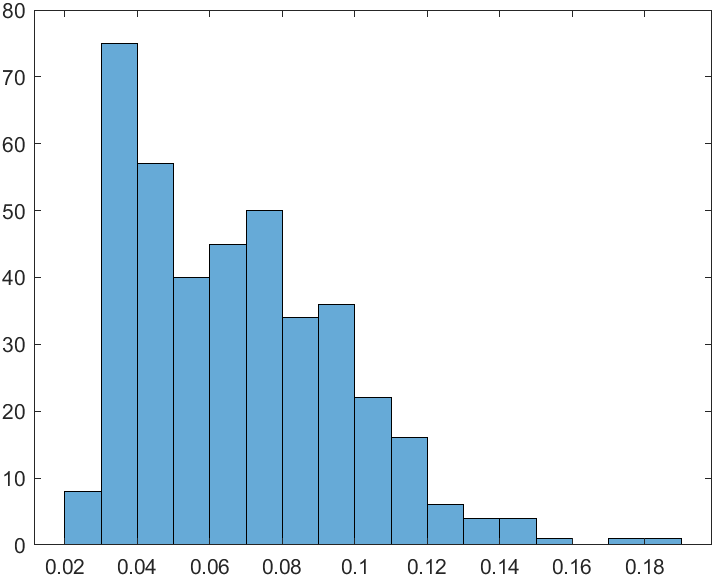
Computational fluid dynamics (CFD) data with 5 input variables is used, and the range of output variables is approximately 0.01 to 0.13. A total of 400 data were used, and the experimental design used LHS.
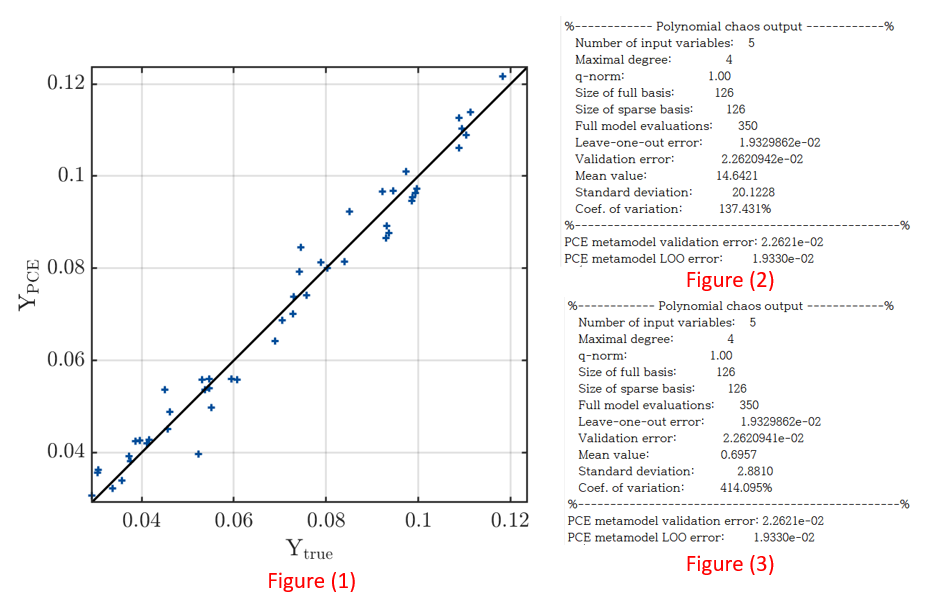
After PCE modeling, the fitting result is the same as figure(1) and the mean is 14.6421 as figure(2). However, considering that the maximum value of the output variable is 0.13, the fitting does not seem to be working properly. In other words, it seems to be overfitting that fits only the given data.
In addition, when the input variable was normalized between 0 and 1 using the normalize (A,‘range’) matlab function, it has the same LOO and validation error as figure(3), but when one input dataset is substituted, the output is 2 Is coming out with a value over. In other words, the fitting is still not working well.
I have some questions in this part.
(1) In my opinion, overfitting occurred due to the small number of data compared to the non-linearity of the data, so the mean value appears to be strange as shown in figure(2). Is this the only way to increase the number of data? Is there any way to prevent overfitting like dropout in neural network?
(2) Determining the degree of PCE is only increasing the order from 1 to finding the optimal degree? Declaring degree as 1:10 keeps converging at 1 degree.
(Actually, the error is the lowest at 4th degree.)
(3) I think that normalization like figure(3) should not change the result. However, in this case, as shown in figure(2) and figure(3), the mean and variance show a large difference. Is this also due to overfitting?
(4) Is there a paper to know the number of required samples according to the number of input variables?
I am looking forward to your advice. I sincerely ask for the advice of researchers.
If you want additional information of my problem please send e-mail to ‘minsu77@yonsei.ac.kr’
Thank you.