Hi everyone.
I would like to ask question about PCE and also the sensitivity analysis. I have 800 output results based on 800 LHS samples with 7 random variables, running in FEM software. Based on the example PCE from existing data, the PCE metamodel was built and had a pretty good validation.
Below is my input data:
%% 4 - PROBABILISTIC INPUT MODEL
InputOpts.Marginals(1).Name = ‘DcRef’;
InputOpts.Marginals(1).Type = ‘Lognormal’;
InputOpts.Marginals(1).Moments = [3e-11 0.6e-11];
InputOpts.Marginals(2).Name = ‘mc’;
InputOpts.Marginals(2).Type = ‘Beta’;
InputOpts.Marginals(2).Parameters = [9.2944 52.6685 0 1];
InputOpts.Marginals(3).Name = ‘Uc’;
InputOpts.Marginals(3).Type = ‘Beta’;
InputOpts.Marginals(3).Parameters = [0.4437 0.1268 32e3 44.6e3];
InputOpts.Marginals(4).Name = ‘depth’;
InputOpts.Marginals(4).Type = ‘Gaussian’;
InputOpts.Marginals(4).Parameters = [0.04 0.01];
InputOpts.Marginals(4).Bounds = [0.01 inf];
InputOpts.Marginals(5).Name = ‘Xr’;
InputOpts.Marginals(5).Type = ‘Gaussian’;
InputOpts.Marginals(5).Parameters = [0.01 0.001];
InputOpts.Marginals(5).Bounds = [0 inf];
InputOpts.Marginals(6).Name = ‘Cfevn’;
InputOpts.Marginals(6).Type = ‘Lognormal’;
InputOpts.Marginals(6).Moments = [7.35 5.145];
% danh cho Cth
InputOpts.Marginals(7).Name = ‘Cth’;
InputOpts.Marginals(7).Type = ‘Gaussian’;
InputOpts.Marginals(7).Parameters = [2 0.4];
% Create an INPUT object based on the specified marginals:
myInput = uq_createInput(InputOpts);
%% 4 - POLYNOMIAL CHAOS EXPANSION (PCE) METAMODEL
% Select PCE as the metamodeling tool:
MetaOpts.Type = ‘Metamodel’;
MetaOpts.MetaType = ‘PCE’;
MetaOpts.Input = myInput;
%%
% Use experimental design loaded from the data files:
MetaOpts.ExpDesign.X = X;
MetaOpts.ExpDesign.Y = Y;
%%
% Set the maximum polynomial degree to 5:
MetaOpts.Degree = 1:5;
%%
% Provide the validation data set to get the validation error:
MetaOpts.ValidationSet.X = Xval;
MetaOpts.ValidationSet.Y = Yval;
%%
% Create the metamodel object and add it to UQLab:
myPCE = uq_createModel(MetaOpts);
%%
% Print a summary of the resulting PCE metamodel:
uq_print(myPCE)
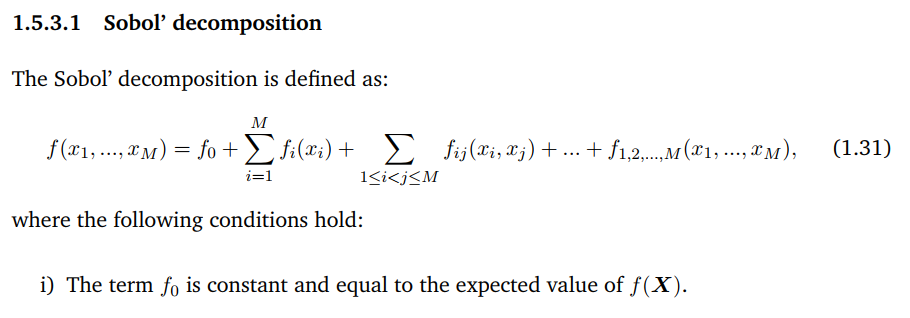
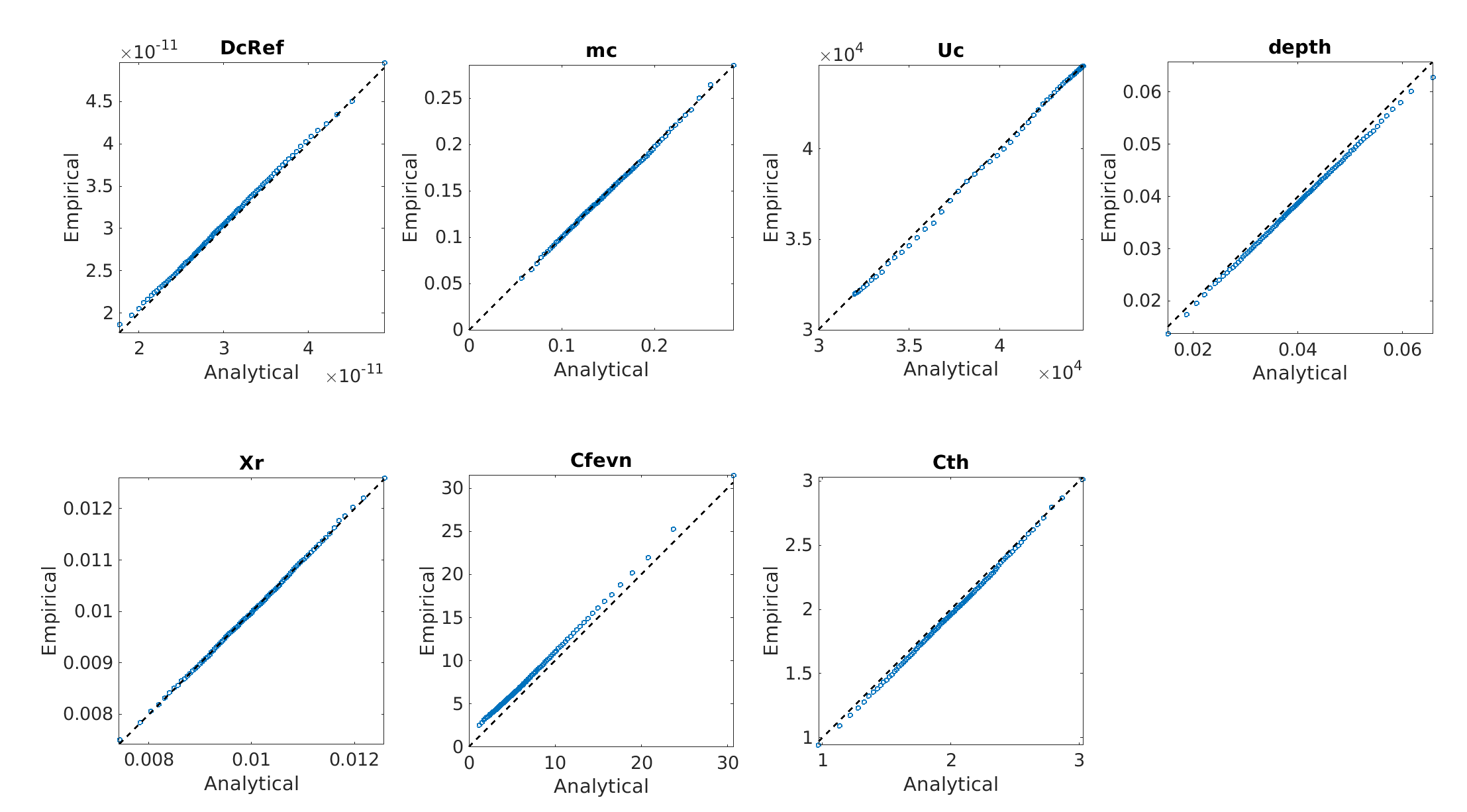
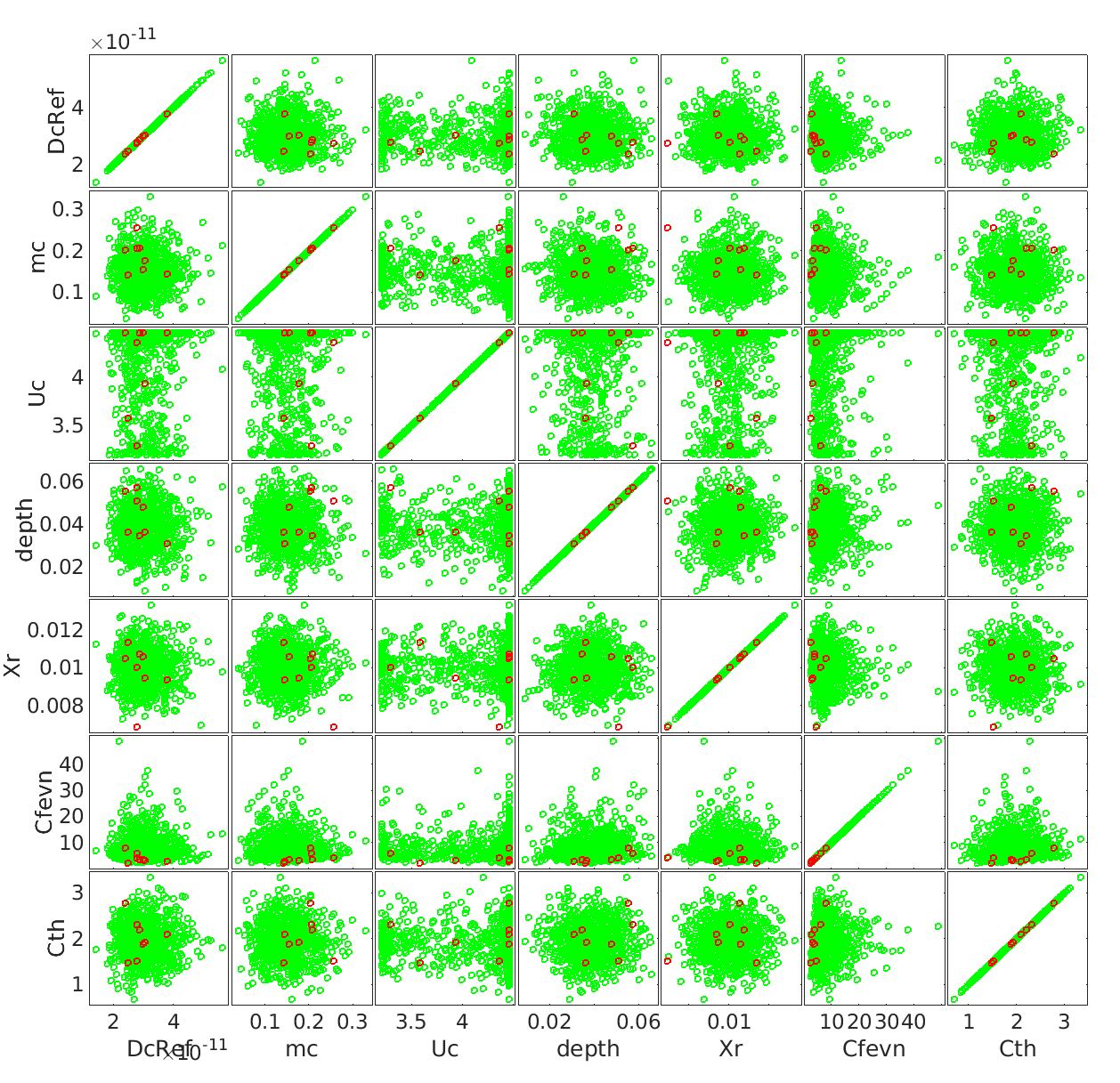
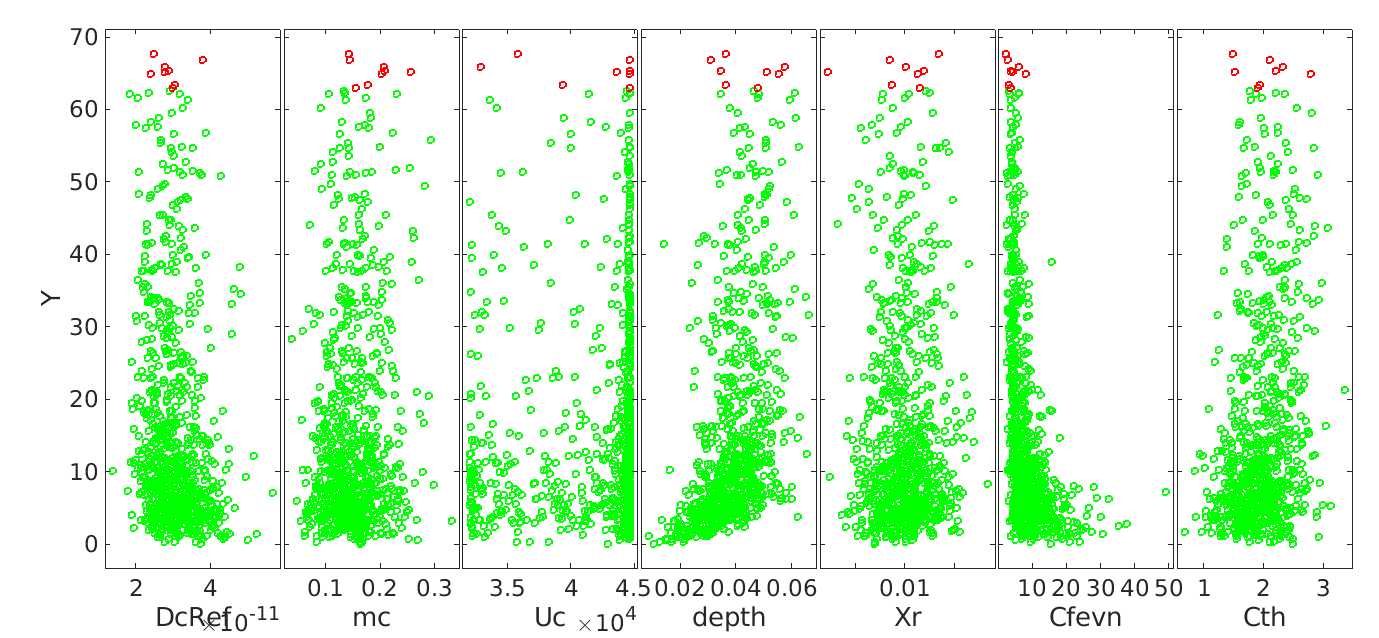
Below is the comparision of True model response and PCE model
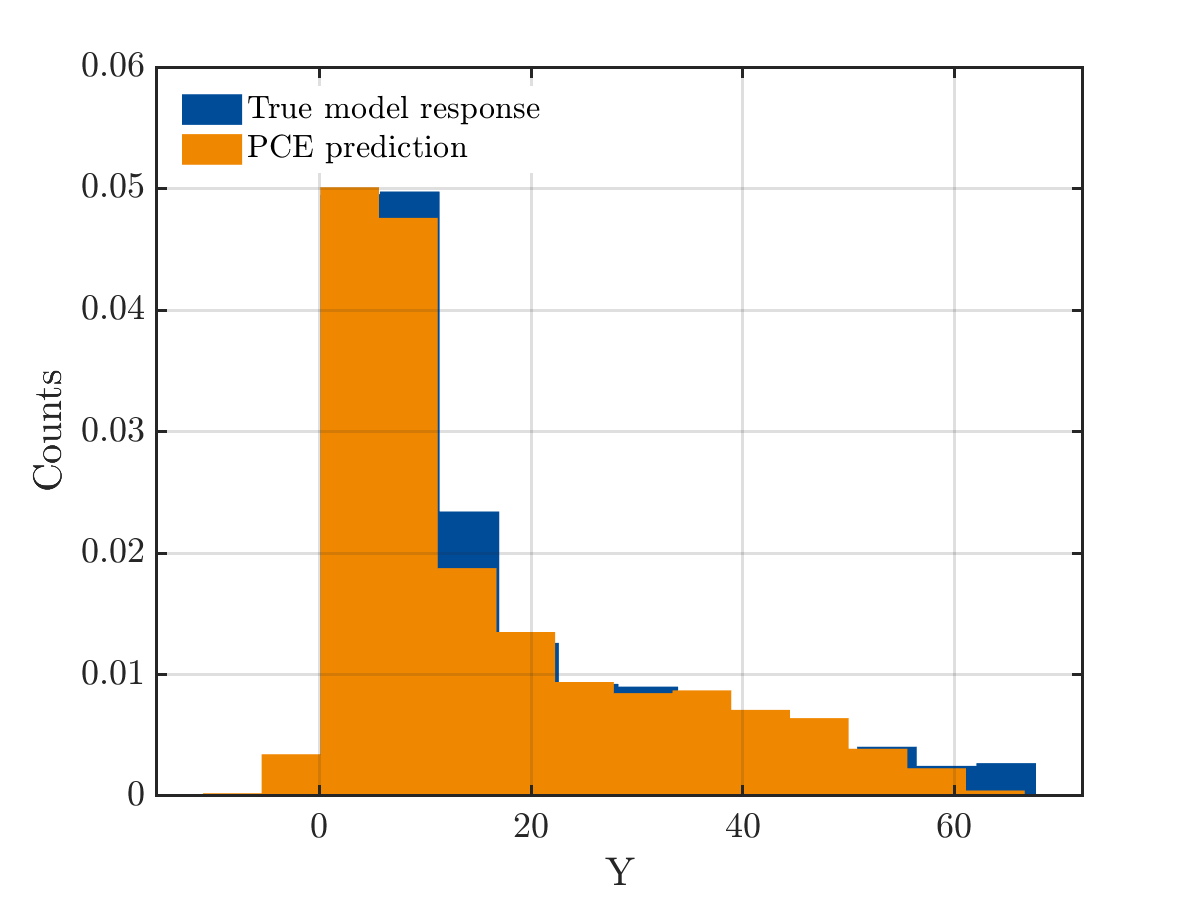
And the true output and PCE output
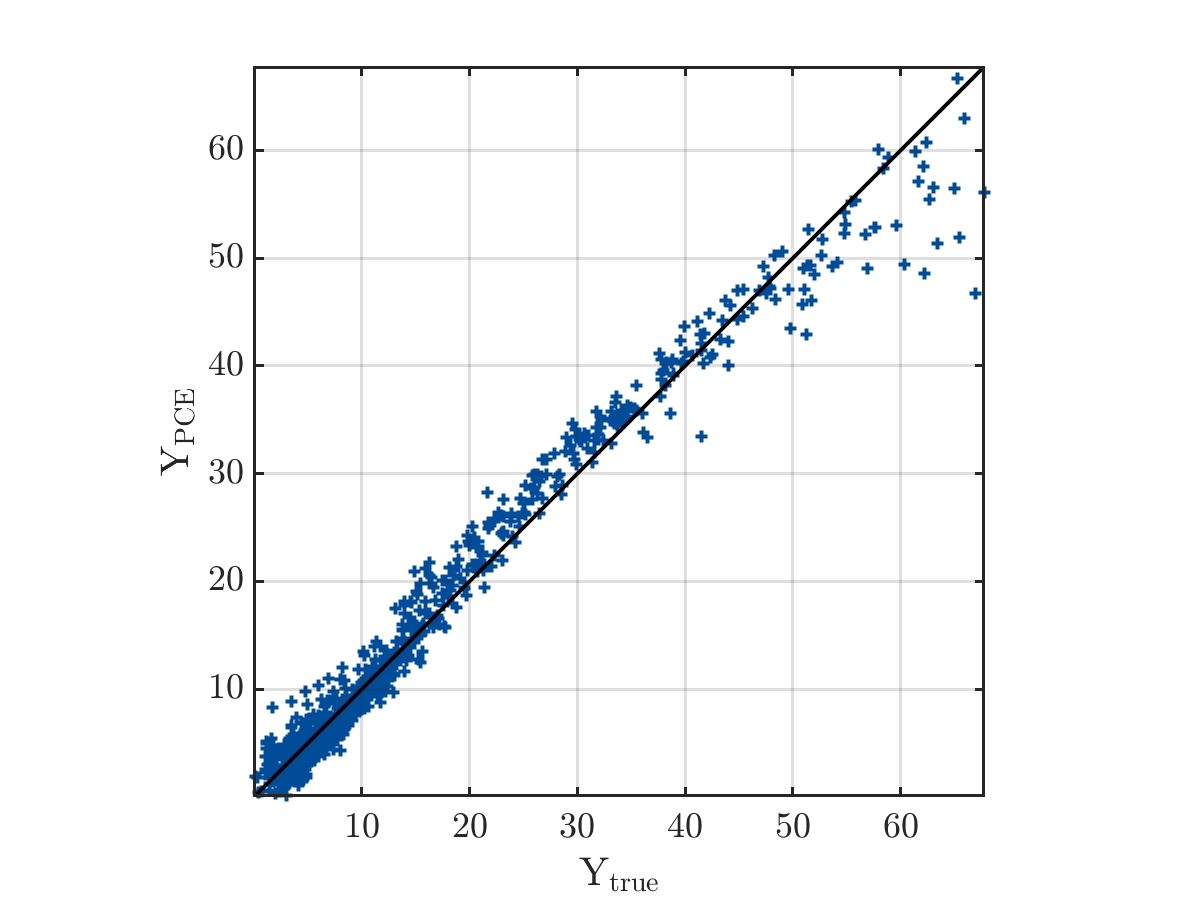
The result from UQlab is shown below:
------------ Polynomial chaos output ------------
Number of input variables: 7
Maximal degree: 3
q-norm: 1.00
Size of full basis: 120
Size of sparse basis: 64
Full model evaluations: 788
Leave-one-out error: 3.3906646e-02
Modified leave-one-out error: 4.5942902e-02
Validation error: 2.6473454e-02
Mean value: 34.6734
Standard deviation: 49.1539
Coef. of variation: 141.762%
--------------------------------------------------
PCE metamodel validation error: 2.6473e-02
PCE metamodel LOO error: 3.3907e-02
The mean value from UQlab is far from the mean value of real data, and as I read in some discussion available in UQlab forum, it does not mean I got the wrong PCE, but it is an arbitrary PCE. Therefore, the mean and the standard deviation of PCE model does not express the real mean and Std of the model. I have checked manually by comparing mean(YPCE) and mean(Yval) and they are the same, so I think the model is still right. My questions are:
- Is there any way to transfer from arbitrary PCE to orthogonal PCE?
- If not, can I do the Sobol’s sensitivity analysis on arbitrary PCE?
- If I have the results of 800 simulations, can I use 800 samples for experimental design and also use the same 800 outputs for validation set?
I really appreciate any support and response from you.
Thank you so much.